Journal:Acta Cryst D:S2059798325007065
From Proteopedia
(Difference between revisions)

| Line 8: | Line 8: | ||
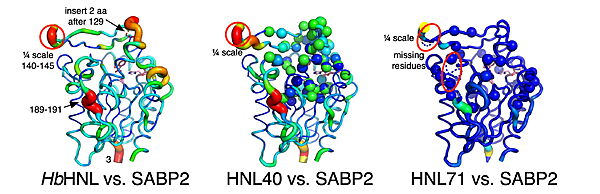
Converting one enzyme into another might seem straightforward—just swap out the residues that directly contact the substrate, and voilà, you have a new catalyst. However, new structural studies reveal that this intuitive approach falls dramatically short. Researchers attempting to transform hydroxynitrile lyase from rubber trees into an esterase discovered that modifying only the 16 residues directly surrounding the substrate produced disappointing catalytic activity. The real breakthrough came when they expanded their engineering to include 40 or 71 mutations, creating variants that not only matched but exceeded the performance of their target esterase. | Converting one enzyme into another might seem straightforward—just swap out the residues that directly contact the substrate, and voilà, you have a new catalyst. However, new structural studies reveal that this intuitive approach falls dramatically short. Researchers attempting to transform hydroxynitrile lyase from rubber trees into an esterase discovered that modifying only the 16 residues directly surrounding the substrate produced disappointing catalytic activity. The real breakthrough came when they expanded their engineering to include 40 or 71 mutations, creating variants that not only matched but exceeded the performance of their target esterase. | ||
| - | [[Image:Calpha_difference_vs_SABP2.jpg|thumb|center|600px|Displacement of Cα atoms (ΔCα) in HbHNL (PDB entry [[1yb6]]), HNL40 (PDB entry [[ | + | [[Image:Calpha_difference_vs_SABP2.jpg|thumb|center|600px|Displacement of Cα atoms (ΔCα) in HbHNL (PDB entry [[1yb6]]), HNL40 (PDB entry [[8sni]]), and HNL71 (PDB entry [[9clr]]) relative to wt SABP2]] |
Revision as of 19:02, 8 August 2025
| |||||||||||
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.