Journal:Acta Cryst D:S2059798323007672
From Proteopedia
(Difference between revisions)

| Line 1: | Line 1: | ||
| - | <StructureSection load='' size='450' side='right' scene='99/992467/Cv/1' caption=''> | + | <StructureSection load='' size='450' side='right' scene='99/992467/Cv/1' caption='View of a typical PTE structure (PDB-ID [[1hzy]])'> |
===The impact of molecular variants, crystallization conditions and the space group on ligand-protein complexes: A case study on bacterial phosphotriesterase=== | ===The impact of molecular variants, crystallization conditions and the space group on ligand-protein complexes: A case study on bacterial phosphotriesterase=== | ||
<big>Orly Dym, Nidhi Aggarwal, Yacov Ashani, Haim Leader, Shira Albeck, Tamar Unger, Shelly Hamer Rogotner, Israel Silman, Dan S. Tawfik and Joel L. Sussman</big> <ref>doi: 10.1107/S2059798323007672</ref> | <big>Orly Dym, Nidhi Aggarwal, Yacov Ashani, Haim Leader, Shira Albeck, Tamar Unger, Shelly Hamer Rogotner, Israel Silman, Dan S. Tawfik and Joel L. Sussman</big> <ref>doi: 10.1107/S2059798323007672</ref> | ||
| Line 5: | Line 5: | ||
[[Image:2023_Dym_Acta_Crys_D_Cover.png| thumb |left|175px|Impact of crystallization conditions on ligands binding to a protein https://journals.iucr.org/d/issues/2023/11/00/nj5323/index.html Go to paper]] | [[Image:2023_Dym_Acta_Crys_D_Cover.png| thumb |left|175px|Impact of crystallization conditions on ligands binding to a protein https://journals.iucr.org/d/issues/2023/11/00/nj5323/index.html Go to paper]] | ||
<b>Molecular Tour</b><br> | <b>Molecular Tour</b><br> | ||
| - | A bacterial phosphotriesterase was employed as an experimental paradigm for examining the effects of multiple factors, such as the molecular constructs, the ligands used during protein expression and purification, crystallization conditions, and space group, on the visualization of molecular complexes of ligands with a target enzyme. In this case, the ligands used were organophosphates that are fragments of the nerve agents and insecticides on which the enzyme acts as a bioscavenger. Twelve crystal structures of various phosphotriesterase constructs obtained by directed evolution were analyzed, with resolutions up to 1.38 Å. Both apo forms and holo forms, complexed with the organophosphate ligands, were studied. Crystals obtained from three different crystallization conditions, crystallized in four space groups, with and without N-terminal tags, were utilized to investigate the impact of these factors on visualizing the organophosphate complexes of the enzyme. The study revealed that tags used for protein expression can lodge in the active site and hinder ligand binding. Furthermore, the space group in which the protein crystallizes can significantly impact the visualization of bound ligands. It was also observed that the crystallization precipitants can compete with, and even preclude, ligand binding, leading to false positives or to incorrect identification of lead drug candidates. One of the co-crystallization conditions enabled us to define the spaces that accommodate the substituents attached to the P atom of several products of organophosphate substrates after detachment of the leaving group. The crystal structures of the complexes of phosphotriesterase with the organophosphate products reveal similar short interaction distances of the two partially charged oxygens of the P-O bonds with the exposed β-Zn<sup>2+</sup> ion and the buried α-Zn<sup>2+</sup>. This suggests that both Zn<sup>2+</sup> ions have a role in stabilizing the transition state for substrate hydrolysis. <scene name='99/992467/Typical_pte/ | + | A bacterial phosphotriesterase was employed as an experimental paradigm for examining the effects of multiple factors, such as the molecular constructs, the ligands used during protein expression and purification, crystallization conditions, and space group, on the visualization of molecular complexes of ligands with a target enzyme. In this case, the ligands used were organophosphates that are fragments of the nerve agents and insecticides on which the enzyme acts as a bioscavenger. Twelve crystal structures of various phosphotriesterase constructs obtained by directed evolution were analyzed, with resolutions up to 1.38 Å. Both apo forms and holo forms, complexed with the organophosphate ligands, were studied. Crystals obtained from three different crystallization conditions, crystallized in four space groups, with and without N-terminal tags, were utilized to investigate the impact of these factors on visualizing the organophosphate complexes of the enzyme. The study revealed that tags used for protein expression can lodge in the active site and hinder ligand binding. Furthermore, the space group in which the protein crystallizes can significantly impact the visualization of bound ligands. It was also observed that the crystallization precipitants can compete with, and even preclude, ligand binding, leading to false positives or to incorrect identification of lead drug candidates. One of the co-crystallization conditions enabled us to define the spaces that accommodate the substituents attached to the P atom of several products of organophosphate substrates after detachment of the leaving group. The crystal structures of the complexes of phosphotriesterase with the organophosphate products reveal similar short interaction distances of the two partially charged oxygens of the P-O bonds with the exposed β-Zn<sup>2+</sup> ion and the buried α-Zn<sup>2+</sup>. This suggests that both Zn<sup>2+</sup> ions have a role in stabilizing the transition state for substrate hydrolysis. <scene name='99/992467/Typical_pte/4'>View of a typical PTE structure</scene> (PDB-ID [[1hzy]]). The (β/α)8 TIM barrel fold is shown as a cartoon, with helices in red, sheets in yellow, coils in green, the α- (buried) and β- (exposed) Zn<sup>2+</sup> ions as magenta spheres, and a single bridging water shown as a cyan sphere. The six residues that bind to the two Zn<sup>2+</sup> ions are shown as ball-and-stick figures, with carbon atoms coloured yellow, nitrogens blue, and oxygens red. The N- and C-terminal residues are labelled N and C, respectively. <scene name='99/992467/Active_site/7'>Close-up view of the active site of the apo-PTE structure</scene>. The buried α-Zn<sup>2+</sup> is directly bound to H55, H57, and D301, while the exposed β-Zn<sup>2+</sup> is bound to H201 and H230. The carbamate functional group bound to K169 interacts with both Zn<sup>2+</sup> ions. Colouring is as in previous scene. |
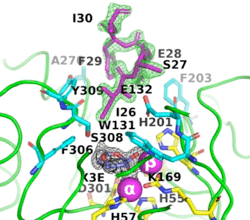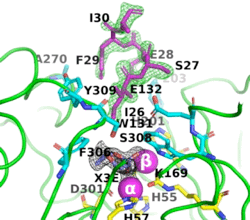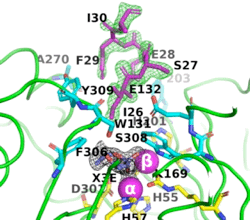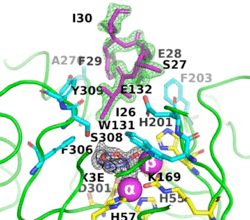
[[Image:Fig_6.gif|left|275px]]<br /><scene name='99/992467/A53_3/3'>Ribbon representation of the A53_3 variant</scene>. It shows the octapeptide tag on subunit B penetrating the active-site region of the symmetry-related subunit A. The tag is shown in magenta. The active-site residues of the symmetrically related chain A are shown in yellow, with those residues within 5 Å of the tag shown in cyan. The cyclic compound, X3E, presumably carried over from the protein expression and purification process, is seen in the active site. The two Zn<sup>+2</sup> ions are shown as magenta spheres. | [[Image:Fig_6.gif|left|275px]]<br /><scene name='99/992467/A53_3/3'>Ribbon representation of the A53_3 variant</scene>. It shows the octapeptide tag on subunit B penetrating the active-site region of the symmetry-related subunit A. The tag is shown in magenta. The active-site residues of the symmetrically related chain A are shown in yellow, with those residues within 5 Å of the tag shown in cyan. The cyclic compound, X3E, presumably carried over from the protein expression and purification process, is seen in the active site. The two Zn<sup>+2</sup> ions are shown as magenta spheres. | ||
Revision as of 09:41, 31 August 2025
| |||||||||||
Proteopedia Page Contributors and Editors (what is this?)
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.