This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
User:Luis E Ramirez-Tapia/Sandbox 1
From Proteopedia
(→This is a placeholder) |
|||
| Line 8: | Line 8: | ||
<!-- | <!-- | ||
| - | + | ||
==What is a Helicase?== | ==What is a Helicase?== | ||
| + | {{STRUCTURE_1jpr | PDB=1jpr | SCENE= }} | ||
Helicases are nucleic acid–dependent ATP-ases that are capable of unwinding DNA [http://en.wikipedia.org/wiki/DNA] or RNA [http://en.wikipedia.org/wiki/RNA] duplex substrates. As a consequence, they play roles in almost every process in cells that involves nucleic acids, including DNA replication and repair, transcription, translation, ribosome synthesis (1) | Helicases are nucleic acid–dependent ATP-ases that are capable of unwinding DNA [http://en.wikipedia.org/wiki/DNA] or RNA [http://en.wikipedia.org/wiki/RNA] duplex substrates. As a consequence, they play roles in almost every process in cells that involves nucleic acids, including DNA replication and repair, transcription, translation, ribosome synthesis (1) | ||
RNA maturation and splicing, and nuclear export processes. Although originally identified by a series of conserved motifs (1), it has become clear more recently that these motifs are actually characteristic of proteins that are able to move directionally along nucleic acid strands, so-called translocases; the helicases themselves being a subgroup of these proteins. Translocase (and helicase) activity is a complex process involving several aspects that together define the mechanisms. | RNA maturation and splicing, and nuclear export processes. Although originally identified by a series of conserved motifs (1), it has become clear more recently that these motifs are actually characteristic of proteins that are able to move directionally along nucleic acid strands, so-called translocases; the helicases themselves being a subgroup of these proteins. Translocase (and helicase) activity is a complex process involving several aspects that together define the mechanisms. | ||
Revision as of 22:36, 3 December 2008
Contents |
This is a placeholder
This is a placeholder text to help you get started in placing a Jmol applet on your page. At any time, click "Show Preview" at the bottom of this page to see how it goes.
Replace the PDB id after the STRUCTURE_ and after PDB= to load and display another structure.
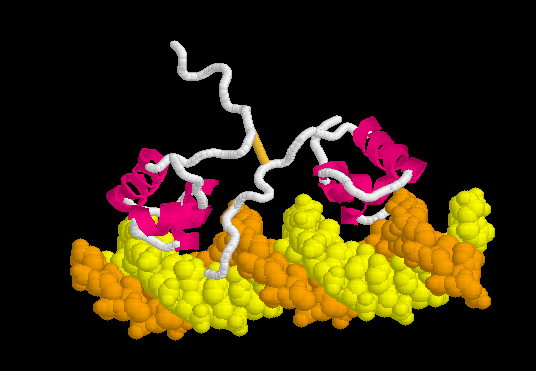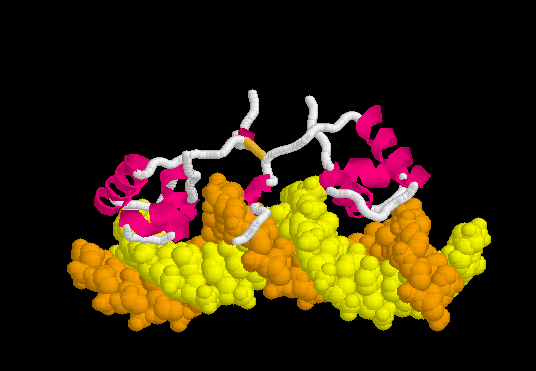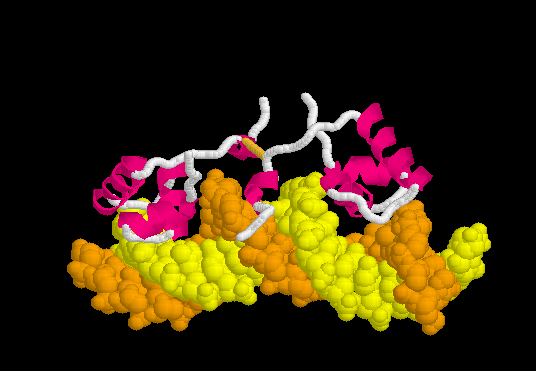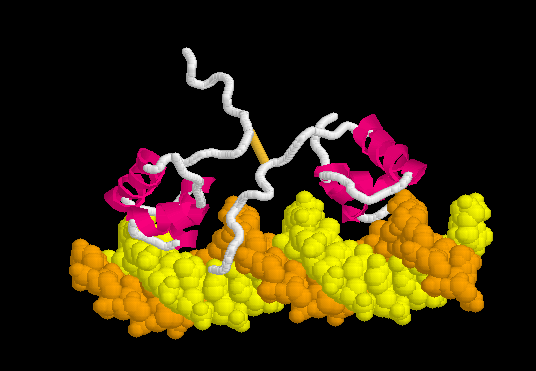
The C-terminal tetramerization helices tether two dimers, and thus the functional form of with two DNA-binding sites.
DNA Binding: Bending the Operator
Non-Specific Binding
|
Lac repressor binds to DNA non-specifically ( derived [1] from 1osl, 20 NMR models), enabling it to slide rapidly along the DNA double helix until it encounters the lac operator sequence. The DNA-binding domain employs a helix-turn-helix motif (Alpha Helices, Turns). During non-specific binding, the hinge region is disordered (indicated by the range of positions of the 20 models), and the DNA double helix is straight. The model shown at right (1osl) has two copies of the DNA-binding domain and hinge region ( to distinguish the chain B hinge). these 20 NMR models simulates thermal motion of the disordered hinge regions.
Specific Binding
Upon recognizing the specific operator sequence, the non-specific binding converts to (derived[1] from 1l1m, 20 NMR models). During this conversion, the hinge region changes from disordered loops to Alpha Helices (), which bind in the minor groove of the DNA. This binding opens the minor groove, bending the DNA double helix. these can be compared with the animation of the non-specific binding.
Morph of Conversion
The can be seen more easily when they are animated smoothly by morphing. (The methods used to create this morph are given in Lac repressor morph methods.) Note the bending of the DNA, with the widening of the central minor groove on the convex aspect. Also note the conversion of the disulfide-bonded hinge region loops to alpha helices. (The displayed secondary structure is calculated for each model in the morph interpolation.)
The specific recognition of the lac operator sequence in the DNA occurs largely though hydrogen bonds. is illustrated in this rendering of the morph. Shown are hydrogen bonds involving Arg22.N-eta2 and Tyr18.OH interacting with DNA base oxygens in the major groove, and Ala53.O interacting with a DNA base nitrogen in the minor groove. (Not all of the relevant hydrogen bonds are shown; see Methods.)
Test:
Animation for Powerpoint® Slides
Here is an animated multi-gif true movie of the above morph, ready to insert into a Powerpoint®[2] slide.
- In Windows, simply drag the movie and drop it into the Powerpoint slide. You can then resize it and position it. The movie should play when you change the View to Slide Show ("project") the slide.
- In Mac OSX, Ctrl-Click on the movie, then Save Image. In Mac Powerpoint, at the desired slide, use the Insert menu (at the top) and select Movie ..., then insert the saved .gif movie file. After inserting the movie, make sure the Toolbox is showing (controlled with an icon-button at the top of the window). Now you can resize and reposition the movie. Click in the movie in the slide to select it. Now, in the Toolbox/Formatting Palette, under Movie, check Loop Until Stopped. Now the movie should play when you change the View to Slide Show ("project") the slide.
Challenge Your Understanding
Here are some questions to challenge your understanding.
- Why does the lac repressor bind to DNA non-specifically?
- When the lac repressor binds non-specifically to DNA, what part of the DNA double helix does it bind to?
- Does DNA have a net charge, and if so, is it negative or positive in aqueous solution at pH 7?
- What kinds of chemical bonds are likely to be involved in non-specific binding of the repressor protein to DNA?
- Does specific binding of lac repressor to DNA disrupt any of the Watson-Crick hydrogen bonds between the base pairs in the DNA strands?
- How do proteins such as the lac repressor recognize specific nucleotide sequences in a DNA double helix?
- What kinds of chemical bonds are involved in specific binding of the repressor protein to DNA?
- Does the lac repressor recognize specific bases in the major or minor grooves of the DNA?
- Why does the lac repressor bend the DNA double helix when it recognizes its specific nucleotide sequence?
Answers are available on request to ![]() . If you would like us to make the answers publically available within Proteopedia, please let us know. When contacting us, please give your full name, your position, institution or school, and location.
. If you would like us to make the answers publically available within Proteopedia, please let us know. When contacting us, please give your full name, your position, institution or school, and location.
Content Attribution
The morphs displayed here were originally prepared by Eric Martz in 2004 for the page Lac Repressor Binding to DNA, within ProteinExplorer.Org.
See Also
- Category: Lac repressor and Category: Lac Repressor, automatically-generated pages that list PDB codes for lac repressor models.
- Morphs where the morph of the lac repressor is used as an example.
References & Notes
- ↑ 1.0 1.1 For these scenes, the 20-model PDB files for 1osl and 1l1m were reduced in size, to avoid exceeding the java memory available to the Jmol applet. All atoms except amino acid alpha carbons and DNA phosphorus atoms were removed using the free program alphac.exe from PDBTools. Secondary structure HELIX records from the original PDB file header were retained. The results are Image:1osl ca.pdb and Image:1l1m ca.pdb.
- ↑ Powerpoint is a registered trademark for a software package licensed by Microsoft Corp..