Sandbox Reserved 1086
From Proteopedia
| This Sandbox is Reserved from 15/04/2015, through 15/06/2015 for use in the course "Protein structure, function and folding" taught by Taru Meri at the University of Helsinki. This reservation includes Sandbox Reserved 1081 through Sandbox Reserved 1090. |
To get started:
More help: Help:Editing |
Contents |
C-cadherin
| |||||||||||
Exploring the Structure
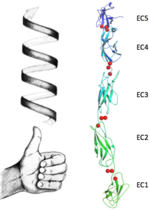
The extracellular domain of C-cadherin is composed of five ectodomains repeated in tandem and linked by short peptide linkers. Each ectodomain (EC) shares the same greek key fold characteristic of immunoglobulin. The interfaces between the EC are calcium binding sites coordinating three ions each through the highly conserved motif Asp-X-Asp, Leu-Asp-Arg-Glu and Asp-X-Asn-Asp-Asn. The binding of calcium it thought to confer rigidity as well as the classical curved shape to the structure and this is why it´s very suggested to be a triggering if not even compulsory step to the adhesion process.
Upon the calcium binding each ectodomain contracts to the neighbour assuming a defined position which leads to the bended structure of the protein. These conformations changes in a sequel of ions form the unique final conformation of this protein. In Fig.1 the bending between EC1 and EC2 is shown in just one dimension, making clear the angle between the two main axes (approximately 22.5°). Fig.2, represents the bending between EC2 and EC3 while concealing this property between first and second ectodomains. Because of this concealing due to the change in the observed direction, the different position of EC3 respect EC2 can be no longer addressed to a bending which takes place on the same direction as the bending between EC1 and EC2 but to a different one. The occurrence of bendings in (at least) two different directions confers a torsional behaviour which goes through all the ectodomains. These particular fashion contributes to the complexity of the protein´s ternary structure, shaping it into a bended left handed helix (Fig.3).
GFP is a beta barrel protein with 11 beta sheets. It is a 26.9kDa protein made up of 238 amino acids. The chromophore, responsible for the fluorescent properties of the protein, is buried inside the beta barrel as part of the central alpha helix passing through the barrel. The forms via spontaneous cyclization and oxidation of three residues in the central alpha helix: -Thr65 (or Ser65)-Tyr66-Gly67. This cyclization and oxidation creates the chromophore's five-membered ring via a new bond between the threonine and the glycine residues [1].
This is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.
This is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.
This is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.
Structural highlights
| 1q5a is a 2 chain structure with sequence from Mus musculus. The March 2008 RCSB PDB Molecule of the Month feature on Cadherin by David S. Goodsell is 10.2210/rcsb_pdb/mom_2008_3. Full crystallographic information is available from OCA. For a guided tour on the structure components use FirstGlance. | |
| Ligands: | , , |
| Related: | 1l3w, 1q55, 1q5b, 1q5c |
| Resources: | FirstGlance, OCA, RCSB, PDBsum |