The interactive Molecular Tour below assumes that you are familiar with the journal article[1].
Introduction
Amyloid fibrils were first assumed to be the agents of Amyloid diseases,including Alzheimer’s,Parkinson’s and the prion conditions.But studies from many laboratories suggest that the reason for this disorder are lower molecular weight entities known as small amyloid oligomers,instead of the associated protein fibrils.Segment of amyloid forming protein makes oligomeric complex which exhibits properties of other amyloid oligomers.They are rich in beta-sheet structure and these oligomer can be identified by a conformational antibody(A11) that binds oligomers but not fibrils,irrespective of sequence of constituent protein.This protein is a chaperone that forms amyloid fibrils.The structure of oligomer shows a cylindrical barrel,made up of six anti-parallel protein strands known as cylindrin.This segment(coloured in black) termed as forms the cylindrin structure.
Molecular Tour
Oligomer forming segment of ABC were identified by inspection of its 3D structure and by applying the Rosetta-Profile algorithm to its sequence.Two segments of high amyloidogenic propensity,with sequences KVKVLG and GDVIEV (where D indicates Asp; E, Glu; G, Gly; I, Ile; K, Lys; and V, Val).
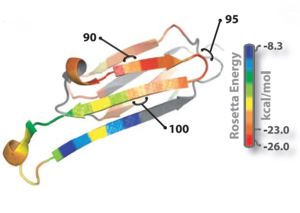
Ribbon diagram of a single subunit of ABC (16), colored by propensity to form amyloid, with red being the highest and blue the lowest propensity. The segment from residue 90 to 100, termed K11V, forms the cylindrin.
The entire 11-residue segment KVKVLGDVIEV forms a in the 3D structure of ABC.The six residue segment GDVIEV ,termed ,forms fibrils and microcrystals.The microcrystals enabled us to determine the atomic structure of G6V, which proved to be a standard class 2 steric zipper, essentially an amyloid-like protofilament.
Amyloid fibrils and oligomer are both formed by the hairpin segment KVKVLGDVIEV.K11V forms fibrils similar to those of protein(ABC) on shaking at elevated temperature and also similar to K11V^V2L(K11V-TR).The fibrils diameter range from 20 to 100 nm in electrom microscope.G6V,K11V,K11V-TR are all convertible to amyloid state,as is their parent protein ABC.Segment K11V,K11V-TR,and a sequence variant with Leu replacing Val at position 2(K11V^2L) forms stable oligomers intermediate in size between monomer and fiber.
K11V and K11V^V2L form hexameric oligomers.K11V oligomer is of 6 chains and K11V-TR oligomer of three tandem chains.
The structure of K11V is a six-stranded antiparallel barrel of cylindrical in shape also called as cylindrin.Each strand of cylindrin is bonded to one neighbouring strand by a strong interface and to a second by a weak interface.The weak interface is formed by eight hydrogen bonds: four from the main chain, two mediated through side-chain interactions, and two through a water bridge.The strong interface is formed by 12 hydrogen bonds and spreads outward at the ends.
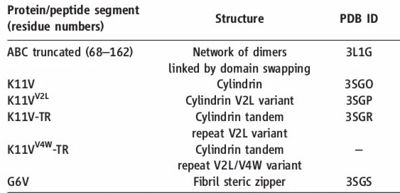
Information about amyloid related oligomers,derived from ABC
PDB'S are
K11V (), K11V-Br2 (), K11V-Br8 (), K11VV2L (), K11V-TR (), and GDVIEV ().
Relevance
Notes & Reference


