User:Yash Patankar/Sandbox 1
From Proteopedia
| |||||||||
| 2cg9, resolution 3.10Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | |||||||||
| Related: | 1a4h, 1ah6, 1ah8, 1am1, 1amw, 1bgq, 1hk7, 1us7, 1usu, 1usv, 2akp, 2brc, 2bre, 2cge | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Contents |
Hsp90
Introduction
Hsp90, a 90 kDa heat-shock protein, is a ubiquitous molecular chaperone responsible for the governing of protein folding and regulation of many eukaryotic signaling pathways in the cell (Ali et al., 2006). It is a non-fibrous protein involved in the activation of many regulatory and signaling client proteins under normal conditions in addition to the refolding of a number of proteins during stress (Ali et al., 2006; Pearl et al., 2008). Hsp90 is one of the most abundant proteins expressed in cells and it has been estimated to constitute upto 2% of the cellular proteins (Dollins et al., 2007). Hsp90 is found in all eukaryotes as well as bacteria with the exception of archae (Chen et al., 2006). Hsp90 has been shown to act as a buffer or capacitor of genetic variation in morphological evolution in Drosophila and other model organisms (Rutherford et al., 2007). Hsp90 is known to be required for the function of protein kinases such as ErbB2m, Cdk4, B-Raf and Akt/protein kinase which are upregualted in cancerous cells (Pearl, 2005).
The function of Hsp90 depends on its ability to hydrolyze ATP after it binds in the active site of the protein and hence, the inhibition of Hsp90 by ATP competitors or non-hydrolyzable ATP analogs leads to the promotion of client degradation.
Structure
The Pearl group (2006) first determined the complete structure of yeast Hsp90 along with the co-chaperone p23 (mammals)/Sba1 (yeast homolog) bound to a non-hydrolyzable ATP analog. This structure was obtained using an Hsp90 mutant (A107N) that was previously shown by the Pearl group to not alter the function of the wild type protein. The PDB id for the protein is 2cg9 and the crystallographic information can be found on OCA______(http://oca.weizmann.ac.il/oca-bin/ocaids).
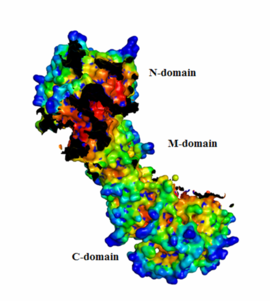
Each Hsp90 monomer consists of an N-teminal domain, a middle (M) domain and a C-terminal domain. The N-terminal domain (a.a. 1-216) is formed by a twisted six stranded β-sheet on one face of the monomer and six α-helices, leading to a two-layer sandwich. The charged linker connecting the N-terminal and M domains consists of poorly conserved and low complexity repeats of amino acids and thus is an obstacle to obtaining crystals showing strong diffraction patterns and was thus omitted from the structure determination process and replaced by an eight residue-loop. The middle domain is subdivided into a large (a.a. 273-409) and small (a.a. 435-525) domain, both of which are α-β-α domains. The C-terminal domain (a.a. 525-709) consists of a three stranded β-sheet a five α-helices. A three-helix coil forms the constitutive dimerization interface for the protomer as shown in figure ______. The residues 678-709 provide the binding sequence for TPR-domain co-chaperones, but they are not crystallized as they are highly disordered (Ali et al., 2006). Sba1/p23 molecules bind to Hsp90 in the junction of the N-terminal domains of the dimer. Figure______ shows a bound Sba1/p23 an Hsp90 monomer.
Conformation of Hsp90
Hsp90 is observed in a distinct “closed” and an “open” conformation. Binding of ATP leads to the transient dimerization (homodimer) of Hsp90 with form the C-terminus domain. The C-terminal domains form a molecular clamp that leads to the dimerization. It is not clear whether the “open” conformation of the dimer exists in vivo as X-ray crystallization studies show that there is defined conformation for Hsp90 in vivo. However, the crystal structures for Hsp90 homologs which lack ATP show distinct conformations as shown in Figure ____________.
- Molecular clamp
The interface between the C-terminal domain and the middle domain flexes upon the binding of ATP, leading to a huge twisted V conformational change that leads to the closure of the domains. The figures show the open as well as the closed state of Hsp90, only for the M and C domains. The structure for all the N, M and C domains in the open conformation is not available for Hsp90, but is available for its homologs such as bacterial HtpG (Pearl et al., 2008).
- N-terminal
The following changes take place in the N-terminal domain upon the binding of ATP:
- The lid segment (a.a. 94-125) swings by ~180o leading to the exposition of a hydrophobic patch on Leu 15 and Leu 18. In addition, the involvement of Gln 14, Thr 95, Ile 96, Ala 97 and Phe 120 leads to an interface between the equivalents on the other monomer (Ali et al., 2006).
- Thr 22, Val 23 and Tyr 24 form hydrophobic interactions with Leu 376 and Leu 378 from the middle domain (Ali et a., 2006).
- Phe 200 makes hydrophobic interactions with Thr 273, Pro 275, Trp 277, Phe 292 and Tyr 344 from the middle domain.
- Amino acid residues from 114 to 120 make polar and hydrophobic interactions with the amino acid residues 372-379.
The interactions between the N and M domains lead to the stabilization of the closed state.
Active site
The constraint from the open state of Hsp90 to its closed state leads to the formation of the active site. Hydrogen bonding between the β-phosphate and the lid segment residues Gly 100 and Val 122 and hydrogen bonding between the γ-phosphate and the lid segment residues Gln 119, Gly 121, Val 122, Gly 123 and Arg 380 occurs due to the movement of the lid. Arg 380, which is part of the M domain, is the only residue that makes a contact wth the ATP outside the N domain. Glu 33 and Arg 380, which are in close proximity of ATP, are thought to play a highly important role in catalysis as their mutations leads to no catalytic activity. Asp 79 is also thought to play a role in the catalytic machinery.
Of note is the fact the interacting residues in the active sites as well as in the hydrophobic and polar interactions of the N and M domains are conserved in the Hsp90 homologs (Ali et al., 2006, Pearl et al., 2008).
HotPatch analysis
ConSurf analysis
Co-chaperone binding
Possible mechanism
Structures for Hsp90 homologs
[[Image: [[Image:Media:Example.jpg
[[Image:Media:Example.jpg ]]]]
]]]]