Sandbox Reserved 460
From Proteopedia
| This Sandbox is Reserved from 13/03/2012, through 01/06/2012 for use in the course "Proteins and Molecular Mechanisms" taught by Robert B. Rose at the North Carolina State University, Raleigh, NC USA. This reservation includes Sandbox Reserved 451 through Sandbox Reserved 500. | |||||||
To get started:
More help: Help:Editing For more help, look at this link: http://www.proteopedia.org/wiki/index.php/Help:Getting_Started_in_Proteopedia
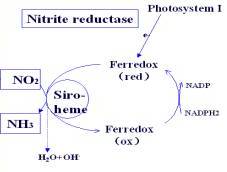
Nitrite ReductaseGeneral InformationNitrogen is an essential element for plant growth and development. Many plants form symbiotic relationships with bacteria and fungi to fix elemental nitrogen from the atmosphere to a deliverable form for the plants to use. Other plants have enzymes that can reduce nitrates in the soil to ammonium in a two step reduction process involving nitrate reductase and nitrite reductase. Without a functional nitrite reductase, the plants accumulate excess nitrites in the cells which are toxic to plants. OverviewNitrite reductase is an enzyme that belongs to the Oxidoreductase family for catalyzing the six-electron reduction of nitrite into ammonia, using the reduced ferredoxin as the electron donor. It is a monomeric protein with molecular mass near 63 kDa characterized by a unique which contains a single [4Fe–4S] cluster and a single siroheme via a bridging sulfur atom from a cysteine residue. The protein has a globular fold consisting of 3 alpha/beta domains with the siroheme-iron sulfur cofactor at the interface of the three domains. The siroheme is surrounded by several ionizable amino acid residues that facilitate the binding and subsequent reduction of nitrite. Nitrite reductase is found in several organisms and different forms that reduce nitrite to various products. The enzyme displayed contains a siroheme and iron-sulfur cluster to reduce nitrite to ammonia in certain plants. It is found in the stromal space of chloroplasts. Several other types of iron-based enzymes in bacteria reduce nitrite to nitrous oxide. There are also several types of Copper Nitrite Reductases found in many different plants and bacteria. MechanismThe electron flow proceeds initially from reduced ferredoxin to the enzyme's [4Fe–4S] cluster and subsequently from the reduced cluster to the siroheme. Since ferredoxin is a one-electron donor, the enzyme must accumulate six electrons in one-electron steps before it can reduce nitrite. Eight protons are also required in the reduction reaction. Reduced ferredoxin binds to the enzyme near the iron-sulfur cluster which becomes reduced. The cluster then transfers an electron to the siroheme. The reduced siroheme binds nitrite. Five more electrons are transferred to siroheme in this way until nitrite is fully reduced.
StructureSecondary StructuresNitrite Reductase has only one chain consisting of 591 residues. There are 33 (33% of chain) and 33 (21% of chain). The enzyme is composed of three alpha/beta which are not as well classified as nitrate reductase. Ligandsis an iron-containing, modified tetrapyrrole similar in structure to both a heme and chlorophyll. It is a heme-like prosthetic group used by nitrite reductase to carry out the six-electron reduction of nitrogen. Nitrite is reduced to ammonia while still bound to siroheme. The siroheme-iron sulfur cofactor is at the interface of the three domains of the enzyme. The siroheme is surrounded by several ionizable amino acid residues that facilitate the binding and subsequent reduction of nitrite. The is a prosthetic group involved in single electron transfer from reduced ferrodoxin to siroheme. Ligand Binding to Active Sites
References
|