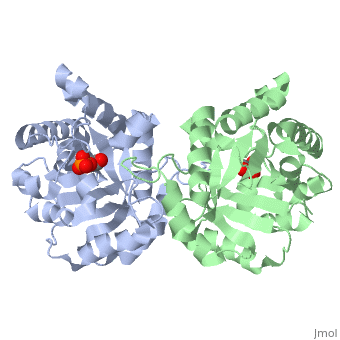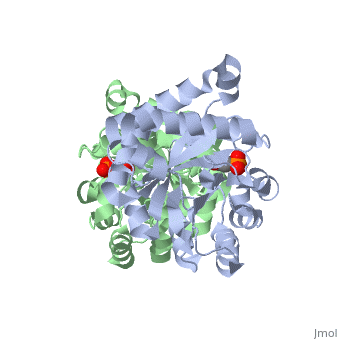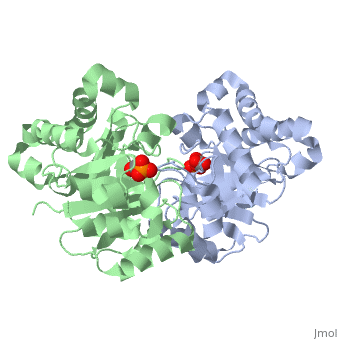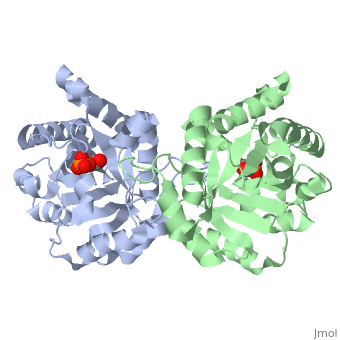Sandbox Reserved 806
From Proteopedia
| This Sandbox is Reserved from Oct 10, 2013, through May 20, 2014 for use in the course "CHEM 410 Biochemistry 1 and 2" taught by Hanna Tims at the Messiah College. This reservation includes Sandbox Reserved 780 through Sandbox Reserved 807. |
To get started:
More help: Help:Editing |
|
Contents |
Introduction
This is , an enzyme in the glycolytic pathway. It is a dimer, but can function as a . It catalyzes the reaction of D-glyceraldehyde 3-phosphate to glycerone phosphate. The ligand that binds to it is called 2-Phosphoglycolic acid (PGA).
3D Structure
The contains alpha helices (gold) and beta sheets (purple). Overall, it forms a beta barrel motif.
was not working.
Residues
Pictured in yellow are the . The are pictured in green.
Solvent Accessibility
are pictured in dark blue. They are mostly located between the beta sheets and alpha helices, in a diagonal line, angled towards the connection point between the two monomers.
Ligand Interaction Site
The ligand---- with the molecule at the location pictured. The that are 4 Å or less from the ligand, are pictured in orange.