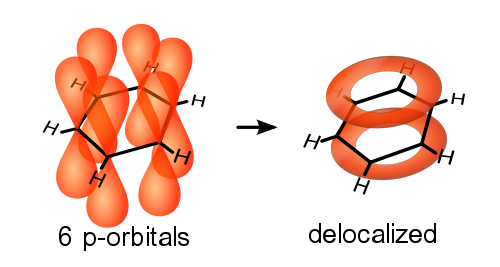|
Nicotinic Acetylcholine Receptor and Nicotine Addiction
Receptor
The nicotinic acetylcholine receptor is a ligand-gated ion channel that responds to the neurotransmitter acetylcholine. It occurs in the central and peripheral nervous systems, and in the neuromuscular junction that controls muscle contraction. It has multiple forms, notably a neuronal type found in the brain, and a muscle type found in neuromuscular junctions. The nicotinic acetylcholine receptor is the basis for nicotine addiction, which kills over 4,000,000 people annually[4]. As stated by Xiu et al.[4]:
"... if nicotine activated ACh receptors found in muscle as potently as it does brain receptors, smoking would cause intolerable and perhaps fatal muscle contractions."[4]
Therefore, the difference between the neuronal type and muscle type of receptors makes nicotine addiction possible, and the chemical basis for the difference is of considerable interest.
Nicotine Cation
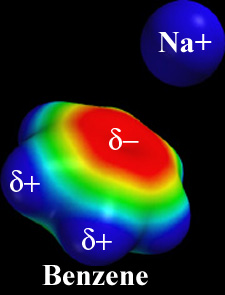
Like acetylcholine, nicotine has a cationic nitrogen when the pH is neutral or acidic. This occurs in nicotine's 5-membered methylpyrrolidine ring (see nicotine structure), which has a pKa of 8.0[5][6].
Cation-Pi Interactions
In 1998, Zhong et al. (with Dougherty)[7] provided evidence that the cation in acetylcholine engages in a cation-pi interaction with Trp149 in the neuronal-type receptor, making an important contribution to the binding affinity. In 2002, Beene et al. (with Dougherty)[8] provided evidence that no such cation-pi interaction is involved when nicotine binds to the muscle-type receptor, thus accounting in part for the lower affinity. In 2009, Xiu et al. (with Dougherty)[4] provided evidence that a strong cation-pi interaction occurs between the cation of nicotine and Trp149 in the neuronal-type receptor. The lower affinity of nicotine for the muscle-type, compared to the neuronal type receptors, is accounted for by differences in cation-pi interaction strength and the absence of a nicotine-receptor hydrogen bond in the former[4].
The crystal structure of a molluscan acetylcholine-binding protein (1uw6) confirms the existence of a cation-pi interaction between the homologous Trp (Trp143) and the nicotine cation (). Also seen is the proposed between nicotine and the main-chain oxygen of the proximal Trp for the rat receptor[4]. The molluscan crystal structure also reveals a remarkable .
Summary
| Ligand-Receptor
| Cation-pi
| Hydrogen bond
|
| Acetylcholine-Neuronal type[7]
| Yes
| (not applicable)
|
| Nicotine-Muscle type[8]
| No
| No
|
| Nicotine-Neuronal type[4]
| Yes
| Yes
|
Examples from Gallivan and Dougherty[1]
- 1gai: a 472-amino acid chain (glucoamylase) with a .
- 1bfg: a 126-amino acid chain (fibroblast growth factor) with an unusually high incidence of cation-pi interactions. Gallivan and Dougherty report 5 energetically significant interactions.
- 2wea: the longest single chain (323 residues) with no energetically significant cation-pi interactions from the original set of 593 proteins originally analyzed.
Example from Zacharias and Dougherty[3]
- 1l8b is a portion of a eukaryotic translation initiation factor that recognizes N7-methylated guanosine on the 5'-end of mRNAs. The .
- 1axi (human growth hormone): Chain B contains an unusual string of three aromatic sidechains separated by, and capped at the ends with, 4 cationic sidechains. (Using "c" for cation and "p" for pi, the chain is "cpcpcpc".) Only the one of these six interactions is deemed energetically insignificant by CaPTURE (Lys at one end). See image here.
- 1bl8 (bacterial potassium channel): There is a for each contact between chains of this homotetramer, but no intrachain cation-pi interactions.
- 2vab (peptide bound to class I histocompatibility protein): The N-terminal Phe of the nonapeptide stacks with Trp 167 of the protein. On either side of the stacked rings are cations (Arg 170, Lys 66), forming the unusual cppc chain RWFK. See image here.
- 1rog (peptide bound to class I histocompatibility protein): Three (of the four) cations in the 9-residue peptide interact with aromatic sidechains in the protein groove. This is a theoretical model.
- 1dlh (peptide bound to class II histocompatibility protein): There are no cation-pi interactions for the 13-residue peptide, despite its containing three lysines and one tyrosine. A number of nearby sidechains that potentially could interact appear to be blocked by other noncovalent bonding interactions, and the peptide lysine sidechains are generally pointing away from the protein.
Additional Related structures
Literature References & Notes
- ↑ 1.0 1.1 1.2 1.3 Gallivan JP, Dougherty DA. Cation-pi interactions in structural biology. Proc Natl Acad Sci U S A. 1999 Aug 17;96(17):9459-64. PMID:10449714
- ↑ Eric Martz thanks Peter Lillya for help writing this paragraph.
- ↑ 3.0 3.1 Zacharias N, Dougherty DA. Cation-pi interactions in ligand recognition and catalysis. Trends Pharmacol Sci. 2002 Jun;23(6):281-7. PMID:12084634
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 4.6 Xiu X, Puskar NL, Shanata JA, Lester HA, Dougherty DA. Nicotine binding to brain receptors requires a strong cation-pi interaction. Nature. 2009 Mar 26;458(7237):534-7. Epub 2009 Mar 1. PMID:19252481 doi:10.1038/nature07768
- ↑ In contrast, the 6-membered pyridine ring has a pKa of 3.1, so is uncharged at neutral pH. The uncharged or free base form of nicotine is more volatile than the charged form. Cigarette manufacturers have long added ammonia to tobacco to increase absorption of nicotine by smokers. See Summerfield, 1999 cited elsewhere for details.
- ↑ Summerfield, J. H. An acid-base chemistry example: conversion of nicotine. J. Chem. Ed. 76:1397-8 (1999).
- ↑ 7.0 7.1 Zhong W, Gallivan JP, Zhang Y, Li L, Lester HA, Dougherty DA. From ab initio quantum mechanics to molecular neurobiology: a cation-pi binding site in the nicotinic receptor. Proc Natl Acad Sci U S A. 1998 Oct 13;95(21):12088-93. PMID:9770444
- ↑ 8.0 8.1 Beene DL, Brandt GS, Zhong W, Zacharias NM, Lester HA, Dougherty DA. Cation-pi interactions in ligand recognition by serotonergic (5-HT3A) and nicotinic acetylcholine receptors: the anomalous binding properties of nicotine. Biochemistry. 2002 Aug 13;41(32):10262-9. PMID:12162741
- ↑ These examples were noted in the documentation for Protein Explorer: see Content Attribution for the original documentation.
Additional Literature
- Dougherty DA. Cation-pi interactions in chemistry and biology: a new view of benzene, Phe, Tyr, and Trp. Science. 1996 Jan 12;271(5246):163-8. PMID:8539615
- Beene DL, Brandt GS, Zhong W, Zacharias NM, Lester HA, Dougherty DA. Cation-pi interactions in ligand recognition by serotonergic (5-HT3A) and nicotinic acetylcholine receptors: the anomalous binding properties of nicotine. Biochemistry. 2002 Aug 13;41(32):10262-9. PMID:12162741
- Gromiha MM, Santhosh C, Ahmad S. Structural analysis of cation-pi interactions in DNA binding proteins. Int J Biol Macromol. 2004 Jun;34(3):203-11. PMID:15225993 doi:http://dx.doi.org/10.1016/j.ijbiomac.2004.04.003
- Crowley PB, Golovin A. Cation-pi interactions in protein-protein interfaces. Proteins. 2005 May 1;59(2):231-9. PMID:15726638 doi:http://dx.doi.org/10.1002/prot.20417
- Gromiha MM, Suwa M. Structural analysis of residues involving cation-pi interactions in different folding types of membrane proteins. Int J Biol Macromol. 2005 Mar;35(1-2):55-62. Epub 2005 Jan 28. PMID:15769516 doi:http://dx.doi.org/10.1016/j.ijbiomac.2004.12.001
- Prajapati RS, Sirajuddin M, Durani V, Sreeramulu S, Varadarajan R. Contribution of cation-pi interactions to protein stability. Biochemistry. 2006 Dec 19;45(50):15000-10. PMID:17154537 doi:http://dx.doi.org/10.1021/bi061275f
- Crowley PB, Golovin A. Cation-pi interactions in protein-protein interfaces. Proteins. 2005 May 1;59(2):231-9. PMID:15726638 doi:http://dx.doi.org/10.1002/prot.20417
- Dougherty DA. Cation-pi interactions involving aromatic amino acids. J Nutr. 2007 Jun;137(6 Suppl 1):1504S-1508S; discussion 1516S-1517S. PMID:17513416
- Wu R, McMahon TB. Investigation of cation-pi interactions in biological systems. J Am Chem Soc. 2008 Sep 24;130(38):12554-5. Epub 2008 Aug 29. PMID:18759391 doi:http://dx.doi.org/10.1021/ja802117s
- Sussman JL, Harel M, Frolow F, Oefner C, Goldman A, Toker L, Silman I. Atomic structure of acetylcholinesterase from Torpedo californica: a prototypic acetylcholine-binding protein. Science. 1991 Aug 23;253(5022):872-9. PMID:1678899
- Harel M, Aharoni A, Gaidukov L, Brumshtein B, Khersonsky O, Meged R, Dvir H, Ravelli RB, McCarthy A, Toker L, Silman I, Sussman JL, Tawfik DS. Structure and evolution of the serum paraoxonase family of detoxifying and anti-atherosclerotic enzymes. Nat Struct Mol Biol. 2004 May;11(5):412-9. Epub 2004 Apr 18. PMID:15098021 doi:10.1038/nsmb767
- Felder C, Jiang HL, Zhu WL, Chen, KX, Silman I, Botti, SA, Sussman JL. Quantum/classical mechanical comparison of cation-π interactions between tetramethylammonium and benzene. J. Phys. Chem. 2001 A 105: 1326-1333. Paper.
- Tan XJ, Zhu WL, Cui M, Luo XM, Gu JD, Silman I, Sussman JL, Jiang HL, Ji RY, Chen KX. Noncovalent interaction or chemical bonding between alkaline earth cations and benzene? A quantum chemistry study with MP2 and density-functional theory methods. Chem. Phys. Lett. 2001 349:113-122. Abstract
- Sussman JL, Silman I. Acetylcholinesterase: structure and use as a model for specific cation-protein interactions. Curr. Opin. Struct. Biol. 1992 2:721-729. Abstract
See Also
Content Attribution
The initial content of this page was adapted, with the permission of Eric Martz, from the Glossary and Introduction, Gallery & Tutorial for Cation-Pi Interactions that accompanies Protein Explorer. Eric Martz is grateful to Peter Lillya for help with chemical terminology and concepts.
|