Sandbox Reserved 1235
From Proteopedia
Cite error: Invalid <ref> tag;
refs with no name must have content
| This Sandbox is Reserved from Jan 17 through June 31, 2017 for use in the course Biochemistry II taught by Jason Telford at the Maryville University, St. Louis, USA. This reservation includes Sandbox Reserved 1225 through Sandbox Reserved 1244. |
To get started:
More help: Help:Editing |
Contents |
Antifreeze protein 1
Papers
as in: the use of JSmol in Proteopedia [1] or to the article describing Jmol [2] to the rescue.
Function
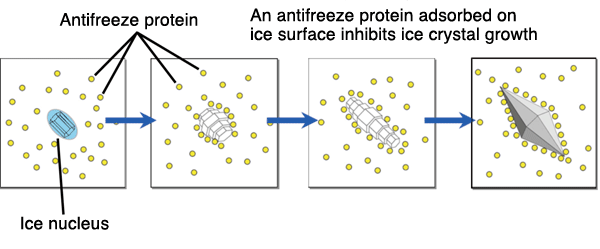
to constrain the process of recrystalization of water as Fishes swim to colder climates , for example like the winter flounder migrating to the northern pole. The molecule is found in many other fish as well that migrate to colder climates such as the shorthorn sculpin, and longhorn sculpin. The function is to depress freezing temperature in order to stop the process of recrystallization to occur.[3] recrystallization is When water begins to freeze, many small crystals form, but then a few small crystals dominate and grow larger and larger, stealing water molecules from the surrounding small crystals. Thus making it manageable for fishes like the winter flounder to swim in colder climates.[4]
Disease
Genetic disease
Mutations DNA Protein
Relevance
Structural highlights
Structure
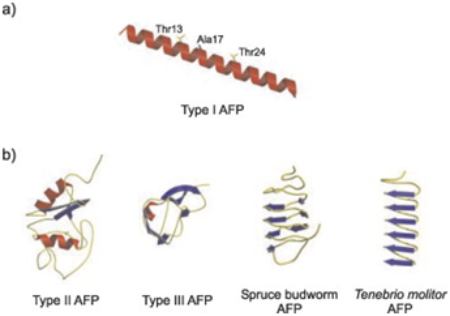
This version of AFP could be recorded as the best documented, for it is the first to have three-dimensional structure determined. it is about 3.3 to 4.5 k Daltons in size. its structure consists of single, long, amphipathic alpha helix. The three dimensional structure consists of three faces: a hydrophobic face, a hydrophilic face, and a Thr-Asx face[8]
|
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ https://www.ncbi.nlm.nih.gov/pubmed/11181960
- ↑ https://pdb101.rcsb.org/motm/120
- ↑ https://en.wikipedia.org/wiki/Antifreeze_protein
- ↑ http://2011.igem.org/Team:KULeuven/Afp
- ↑ http://amazingbiotech.blogspot.com/2014/02/antifreeze-proteins.html
- ↑ http://www.fasebj.org/content/4/8/2460.short