Eukaryotic DNA topoisomerase I (topo I) is a protein that reduces the strain from the supercoils that are caused during transcription and translation[1]. There are two types of topoisomerases. Type 1 topoisomerases are monomeric and break one strand of DNA[2]. Type 2 topoisomerases are dimeric, meaning that they made up of two units and break both strands of the DNA helix[2]. They are able to pass another part of the duplex through the cut, and close the cut using ATP[1].
Structure
Human topo 1 is composed of 765 amino acids [2]. The enzyme consist of 4 regions which are the NH2-terminal, core, linker, and COOH-terminal domains[2]. The NH2-terminal is approximately 210 residues long, it is highly charged, disordered, and contains few hydrophobic amino acids[2]. The COOH-terminal domain is made up of residues 713 to 765 and contains the important amino aside Tyrosine 223[2]. The location of the active site is at this amino acid[2].
Active Site
Topo 1 reduces stress in DNA by causing a transient single strand nick in the the DNA helix[1]. This nick enables the cut to rotate around its intact complement, thus eliminating proximal supercoils[1].
The active site of Topo 1 is catalytic and it is the location where the nicking or cutting occurs[2]. The nicking occurs from the trans-esterification of Tyr-723 at a DNA phophodiester bond forming a 3'-phosphotyrosine covalent enzyme–DNA complex [1]. After the DNA is relaxed, the covalent intermediate is reversed when the released 5'-OH of the broken strand reattacks the phosphotyrosine intermediate in a second transesterification reaction[1].
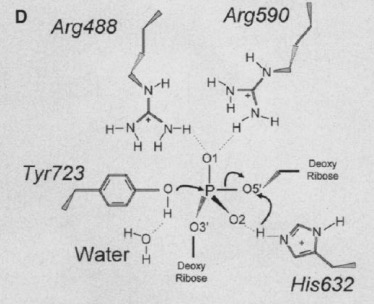 [3]
[3]
Relevance
Many anticancer drugs target topo 1 enzymes.
Mutations
This is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.

