User:Matheus Andrade Bettiol/Sandbox 1
From Proteopedia
==rhoA==
RhoA (Ras homology gene family member A) is a protein of the small GTPase family. It can be in two conformations, and therefore active, or and consequently inactive. Three factors regulate these two states: 1. GEF (Guanine nucleotide exchange factors): promotes the exchange of GDP for GTP, activating RhoA 2. GAP (GTPase activating proteins): accelerates the hydrolysis of GTP, inhibiting RhoA 3. GDI (Guanine nucleotide dissociation inhibitor): translocates the membrane GTPase, sequestering it to the cytosol, also inhibiting RhoA
You may include any references to papers as in: the use of JSmol in Proteopedia [1] or to the article describing Jmol [2] to the rescue.
FunctionThis protein is an important molecular switch for activities associated with the cytoskeleton and the immune system. In relation to the cytoskeleton, it is a regulator or the cell migration, contributing to cell retraction through the ROCK (Rho-associated protein kinase) and LIMK (LIM kinase) pathway, which leads to contraction of acto-myosin II and actin polymerization. In addition, it is relevant for the assembly of occlusive junctions that seal the epithelium in selective permeability barriers, such as in the intestine. When it comes to the immune system, it is essential for the presentation of antigens and formation of immune synapses between the dendritic cell and the T lymphocyte. Not only that, but it also contributes to the recruitment and phagocytosis activity of neutrophils, macrophages and dendritic cells. DiseaseMutations in RHOA have been linked to a predisposition to autoimmune diseases and cancer progression. Additionally, RhoA signaling is possibly involved in the pathogenesis of neurodegenerative diseases, including Parkinson’s disease (PD), Alzheimer’s disease (AD), Huntington’s disease (HD), and amyotrophic lateral sclerosis (ALS). This may be related to the implication of Rho GTPases in brain development since these neurodegenerative diseases present an abnormal accumulation of misfolded peptides. One of these proteins could be RhoA. In addition, different bacteria use a pathogenic strategy of inactivating RhoA through their toxins, which make post-translational modifications in the switch I region. The bacteria and their respective toxins are: - Vibrio parahaemolyticus: VopS (adenylation of Thr 37) - Histophilus somni: IbpA (adenylation of Try 34) - Clostridium botulinum: C3 (ADP-ribosylation of Asn 41) - Clostridium difficile: TcdB/A (glycosylation of Thr 37) - Burkholderia cenocepacia: deamidation of Asn 41
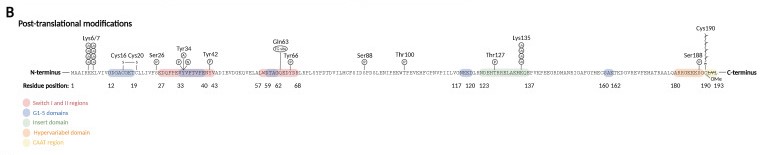
StructureRhoA consists of several domains, each with its specific function: GTPase Domain: This domain is responsible for binding and hydrolyzing . It regulates the activation and inactivation of RhoA by switching between its active (GTP-bound) and inactive (GDP-bound) states. Rho Insert: This is a unique sequence insertion found within the GTPase domain of RhoA. It plays a role in the regulation and interaction of RhoA with other proteins. Switch I and Switch II: These are two regions within the GTPase domain that undergo conformational changes upon GTP binding. Their conformations dictate the ability of RhoA to interact with downstream effector proteins. Insertion/Deletion Sites: RhoA contains four insertion or deletion sites that contribute to the overall structure of the protein. These sites are characteristic of many GTPases in the Rho family and may play a role in protein-protein interactions. C-terminal Prenylation Site: The C-terminal region of RhoA undergoes prenylation, a post-translational modification where a prenyl lipid group (such as farnesyl or geranylgeranyl) is attached. Prenylation allows RhoA to anchor to cell membranes, facilitating its localization and interaction with membrane-associated proteins. Post-Translational ModificationsPrenylation: the activation of Rho GTPases require membrane binding, which is necessary for the interaction with membranous GEFs. The membrane association requires C-terminal prenylation, which involves the addition of a geranylgeranyl (20-carbon chain) to Cys190 in the CAAX motif. Phosphorylation: can alter the subcellular localization of RhoA when occurs close to C-terminal lipid modifications. On the other hand, phosphorylation of the G-domain affects GTP/GDP cycling and the interaction with effector proteins. Oxidation: RhoA can be oxidized on Cys16 and Cys20 (G1 domain), generating a disulfide bond that prevents guanine binding and GEF association, inactivating RhoA. However, if Tyr42 is phosphorylated, serving as a binding site for GEF, oxidation on Cys16/20 reduces the affinity of RhoA for GDI and increases the association with GEF, leading to RhoA activation. Nitration: nitration on RhoA's Tyr34 (switch I region) introduces a negative charge that modifies the protein structure and leads to a faster GDP release and GTP reload, increasing RhoA activity. Adenylation: adenylation on Tyr34 (switch I region) leads to RhoA inhibition. Ubiquitination: target the protein for degradation by the proteasome. RhoA is ubiquitylated by E3 ubiquitin ligase complexes, that ubiquitinate either active RhoA on Lys6 and Lys7, inactive RhoA, or both states on Lys135. Image from the article "RhoA Signaling in Neurodegenerative Diseases", by Sissel Ida Schmidt. This is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.
| ||||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644