Green Fluorescent Protein
From Proteopedia
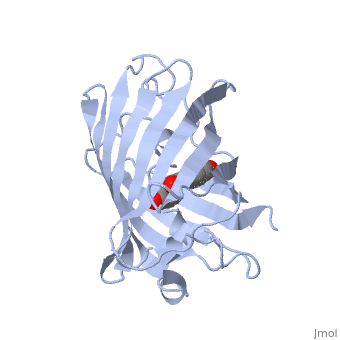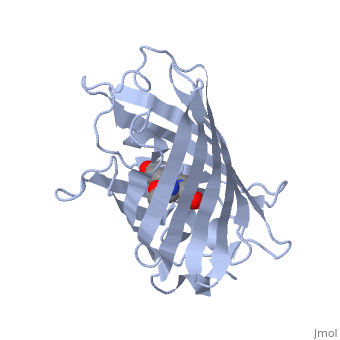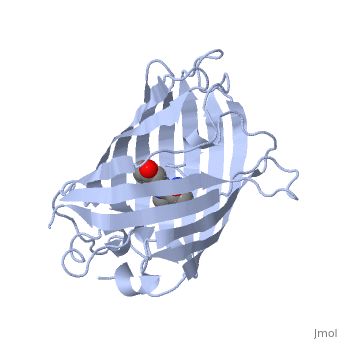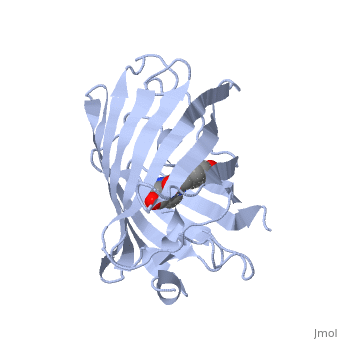
Green Flourscent Protein
with flourescence-conferring chromophore
at center of the barrel-like structure
Contents |
Background
Osamu Shimomura, Martin Chalfie and Roger Y. Tsien shared the 2008 Nobel Prize in Chemistry for their for the discovery and development of the green fluorescent protein, GFP.
GFP is small protein (21 kDa) that naturally occurs in the jellyfish Aequorea victoria and does not require cofactors to become fluorescent. The chromophore at the center of the structure that is responsible for its fluorescence is formed
spontaneously from a tri-peptide motif in the primary structure of GFP. Being small and facile has lead to its use by investigators as a tool to examine a multitude of processes in many organisms; often in such research, GFP is fused to other proteins genetically.
Structure
| |||||||
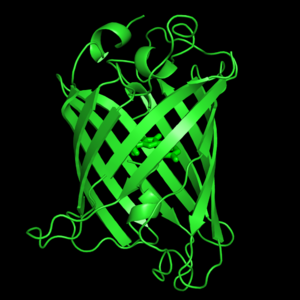
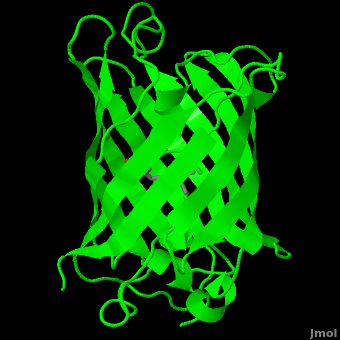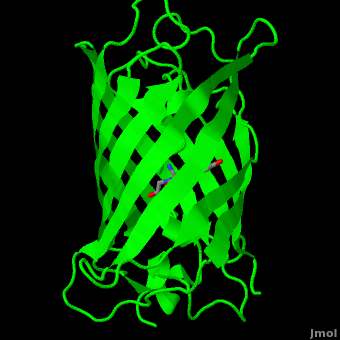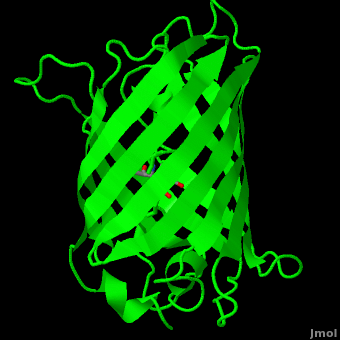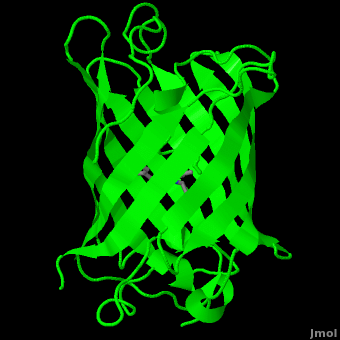
The crystal structure of GFP [1][2] is an eleven-stranded anti-parallel beta-barrel, threaded by an alpha-helix, running up along the axis of the cylinder. In the structure is colored by secondary structure(Alpha Helices and Beta Strands ) to better show the eleven strands and the helices.
.
:
The chromophore is in the distorted alpha-helix that runs along the axis of the can, close to the center of the can-like cylinder.
The chromophore is formed from the tripeptide motif Serine65-Tyrosine66-Glycine67 after translation and folding of GFP. As the GFP protein folds into its native conformation, these three amino acids are forced into a sharp turn, greatly favoring a nucleophilic attack of the amide of Glycine67 on the carbonyl of Serine65, leading to imidazolinone formation by cyclization and dehydration. At this point, GFP is not fluorescent; however, in the presence of molecular oxygen, the α–β bond of Tyrosine66 is subsequently dehydrogenated into conjugation with the imidazolinone, which results in maturation of the GFP chromophore to its fluorescent form [3][4].
The chromophore .
Reference for the Structure
- Ormo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996 Sep 6;273(5280):1392-5. PMID:8703075
Related Structures and Topics
- 1ema Aequorea victoria Green Fluorescent Protein
- 1gfl Aequorea victoria Green Fluorescent Protein
- 1b9c Aequorea victoria Green Fluorescent Protein Mutant F99s, M153t And V163a
- GFP featured at the Molecule of the Month series of tutorials by David Goodsell.
Notes and Literature References
- ↑ Ormo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996 Sep 6;273(5280):1392-5. PMID:8703075
- ↑ Yang F, Moss LG, Phillips GN Jr. The molecular structure of green fluorescent protein. Nat Biotechnol. 1996 Oct;14(10):1246-51. PMID:9631087 doi:10.1038/nbt1096-1246
- ↑ Heim R, Prasher DC, Tsien RY. Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc Natl Acad Sci U S A. 1994 Dec 20;91(26):12501-4. PMID:7809066
- ↑ Cubitt AB, Heim R, Adams SR, Boyd AE, Gross LA, Tsien RY. Understanding, improving and using green fluorescent proteins. Trends Biochem Sci. 1995 Nov;20(11):448-55. PMID:8578587
Additional Literature and Resources
- Tsien RY. The green fluorescent protein. Annu Rev Biochem. 1998;67:509-44. PMID:9759496 doi:10.1146/annurev.biochem.67.1.509
- GFP featured at the Molecule of the Month series of tutorials by David Goodsell.
- The GFP site by Marc Zimmer, Ph. D., at Connecticut College, who authored Glowing Genes: A Revolution In Biotechnology.
Green fluorescent protein (GFP) is a bioluminescent polypeptide consisting of 238 residues isolated from the body of Aequorea victoria jellyfish.[1] GFP converts the blue chemiluminescent of aequorin in the jellyfish into green fluorescent light.[2] In remains unclear why these jellyfish use fluorescence, why green is better than blue, or why they produce a separate protein for green fluorescence as opposed to simply mutating the present aequorin to shift its wavelength,[3] but in the laboratory, GFP can be incorporated into a variety of biological systems in order to function as a marker protein. Since its discovery in 1962, GFP has become a significant contributor to the research of monitoring gene expression, localization, mobility, traffic, interactions between various membrane and cytoplasmic proteins, as well as many others.[4]
History
Aequorea victoria was first discovered and investigated for its bioluminescence by Frank Johnson, who invited Osamu Shimomura to work with him in on a small island not far from British Columbia, where the jellyfish is abundant.[5] Found off the west coast of the United States between British Columbia and central California,[6] the jellyfish was considered a local phenomenon as it would drift in and out of the harbors.[5]
Shimomura was originally looking only to isolate the blue luminescent protein of Aequorea victoria, traditionally thought to be luciferase, but it would soon become apparent that the glow was in fact due to aequorin, a substance related, but slightly varying from luciferase.[4][5] However, the light emitted from aequorin still differed from the light emitted from the wild jellyfish. This quandary led to the discovery of the green fluorescent protein responsible for this disparity, but sufficient amounts of the protein could not be collected for study until 1979. The journey to discover the nature of GFP had begun.[5]
In the 1990’s, Douglas Prasher, Frank Predergast, and co-workers successfully cloned the gene that encoded for GFP. Martin Chalfie further pursued this line of work and was eventually able to express GFP in heterologous systems such as E. coli and C. elegans. Chalfie’s research provided the first evidence that GFP was unique as it did not require the presence of any exogenous substance or cofactor for fluorescence.[4] The lack for the need for a cofactor proved that the cloned GFP gene contained all the information necessary for posttranslational synthesis of the chromophore. [3]
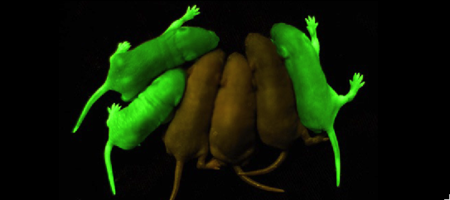
Roger Tsien and co-workers were intrigued by the absence of a necessary cofactor and began to research the structure of GFP and how it relates to its fluorescence. They discovered that a helix within the beta barrel structure of GFP actually contained a fluorophore responsible for fluorescence. In researching its structure, they were able to develop GFP derivatives with improved fluorescence and photo-stability. Shimomura, Chalfie, and Tsien were each recognized for their work involving GFP with the Nobel Prize in 2008.[4] In the time since the work of these three researchers, GFP has been successfully expressed and utilized in bacteria, yeast, slime mold, plants, drosophila fruit flies, zebra-fish, and mammalian cells.[2] Below, mice have had GFP inserted into their genomes for studies in neurology.
Structure
Primary & Secondary Structure
| |||||||||
| 1ema, resolution 1.90Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Non-Standard Residues: | , | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Green fluorescent protein () is a 21 kDa protein consisting of 238 residues strung together to form a
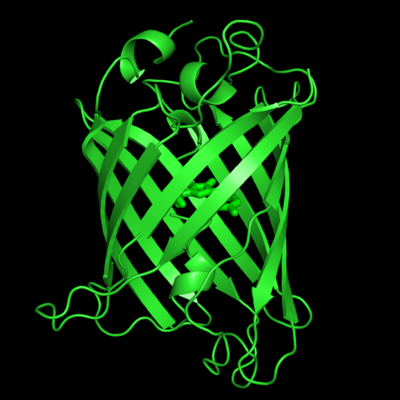
of five α-helices and one eleven-stranded β-pleated sheet,[1] where each strand contains nine to thirteen residues each.[7] (To view the primary and secondary structure of GFP, go to [www.ebi.aci.uk].) These β-strands display an almost “seamless symmetry” in which only two of the strands vary in structural content.[8] This β-sheet conforms itself through regular hydrogen bonding into a β-barrel.[2] In GFP, the structure is so regular that of water molecules (red) can be seen following the structure of the barrel.[8] Together with the α-helices at either end of the molecule, a nearly perfect cylinder is produced, 42Å long and 24Å in diameter,[7] creating what is referred to as a “β-can” formation.[8] The short helical segments at either end of the cylinder form “caps” to further protect the interior of the β-barrel.[8] Overall stability is maintained by this β-can structure, helping to resist unfolding from heat and other denaturants.[2]
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Wayne Decatur, Karsten Theis, Eran Hodis, Laura Carbone, Karl Oberholser, Mark Hoelzer, Joel L. Sussman, Alexander Berchansky, Jaime Prilusky, Marius Mihasan, Joseph M. Steinberger, David Canner