Sandbox 645
From Proteopedia
HIV-1 Protease
Human Immunodeficiency Virus-1 Protease
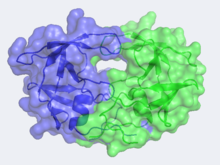
IntroductionHIV -1 protease (HIV PR ) is a retroviral aspartyl protein that is derived from HIV-1, a lentivirus that is best characterized for its ability to lower host immunity by infecting CD4+ T lymphocytes, macrophages, and dendritic cells. Unlike most members of aspartyl protease class, which generally exist as two domain monomers, HIV protease is a dimmer with two identical subunits that are comprised of 99 amino acids. The HIV PR, together with single stranded RNA (ssRNA), reverse transcriptase, integrase, and other viral factors, is found inside the HIV-1 virion. As an important viral protein, it plays a crucial role in successful viral propagation.
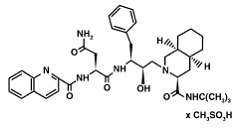
Structure & FunctionMechanismApplications & ResearchWhen a HIV virus infects an organism it tends to multiply within the body’s cells. The virus is then released to infect other cells. In this manner, the infection of HIV infects the newly made cells of the body. While the viruses are produced, proteins and enzymes used to manufacture the DNA in addition to other components of the virus are made. In this case, protease is that enzyme that is needed to bring the structural and enzymes of the virus together. Protease drugs are what could inhibit this virus. HIV Drugs: 1) Saquinavir (Invirase) is known to be one of the first FDA approved protease inhibitor for HIV treatment. This usually occurs by HIV protease binding an active site tunnel tightly, which will prevent polyproteins from also binding. HIV’s chemical structure has the ability to mimic the tetrahedral intermediate of the hydrolytic reaction to interact strong with the catalytic Asp residues. Knowing that, Saquinavir is an uncleavable ligand by studying its similar conformational changes in binding saquinavir or a polypeptide. ((http://www.rxlist.com/invirase-drug.htm))))
((http://www.ncbi.nlm.nih.gov/pubmedhealth/PMH0001007/))
APPLICATIONS: Design of a HIV-1 Protease inhibitor - Free-energy parameterization of enzyme-inhibitor binding. Based on the crystal structure data and protein receptor ligand complexes studied, interatomic interactions that work on burying atoms and find the statistical preference for amino acid pairs. A free energy model of the receptor-ligand is formulated and helps in showing the interfacial interactions. The interaction strength of this model has a reliability of ±1.5 kcal/mol, which reveals the importance of atomic interaction to stabilize the receptor-ligand interface. The analysis of a binding motif of HIV-1 protease inhibitor complex shows the important contacts instead of the set of atoms.
References | ||||||||||||