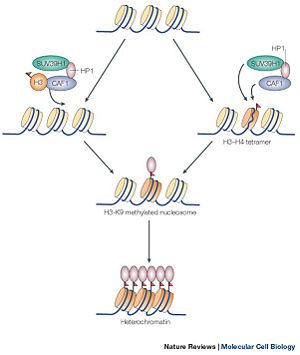
 Figure 1 showing the methylation of the histone protein and the subsequent formation of heterochromatin
Structure
Chromodomain Structure
Crystal structure shows two main parts to the protein, but as a whole three independent molecules. There is a chromodomain and SET domain. The chromodomain starts at the N-terminus of the enzyme and continues toward the C-terminus, where the SET catalytic domain is located. The chromodomain length is around 44-106 amino acids long, which forms three antiparellel beta sheets. The lengths for each of three beta sheets are 45-53, 58-64 and 73-76 amino acids long for beta 1, beta 2 and beta 3, respectively. These three beta sheets form the the chromodomain.
SET Structure
The catylatic domain is consisted of a alpha doible helix. The double helix is located on the C-terminus end of the portein. The residue consisting the catylatic domain is about 82-100 amino acids long. In addition to the two main domains to the enzyme, there is an essential hydrophobic core which is very similar to other chromodomain proteins. The hydrophobic core is made up several residues. These reisdues are V45, L48, Y60, V62, W64, L80, I85 and L86 (each letter represents an amino acids). The similarity between the chromodomain of SUV39h1 and chromodomains of other enzymes is very similar. SUV39h1 has been shown to very similar to the chromoddomain of MPP8 and HP1, showing a conservation in chromodomain structure. although the chromodomain structure is very similar, there is a slight difference with the catylitic domain being longer. In addition to the catyltic domain of SUV39H1 being longer, The enzyme lacks a F34 aromatic cage, which was originally thought to be essential for recognizing exposed lysine or argon residue. However, residues of form a loop which binds to the exposed rsidues of the substrate, showing that it is not a conserved feature in the chromodomain family of enzymes.
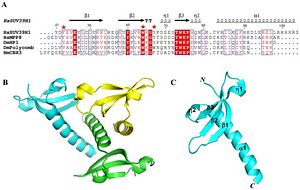 Figure 2 shows the residue and tertiary and Quaternary structure SUV39H1
Function
SET Function
There are two types of histone methyltransferases: lysine specific and argine specific; each are residues which the enzyme transfers a mathyl group to. Within the lysine specific, there are further two main types: SET (Su(var)3-9, Enhancer of Zeste, Trithorax) and non-SET domains. These domains are an essential part of the enzyme because the main catalytic occurs in SET domain of the methyltransferase. SUV39h1 is considered to have a lysine specific SET domain which catalyzes the methyltransferase. In the nucleosomes, there are four different proteins; each consists of two copies which make the entire nucleosome. The mechanism by which SUV39H1 methylates the histone proteins is also known. A nearby tyrosine residue in the histone protein creates a strong nucleophile by deprotonating a nearby lysine residue in the same histone protein. The lysine residue is a very strong nucleophile which attacks the sulfur atom of the cofactor S-Adensyl methoinine (SAM)and extracts a methyl group from the cofactor. subsequently, the attack methylates the histone protein. The cofactor is very important because it is the source of the methyl group.
Chromodomain function
---
Clinical relevence
A mutation in SUV39H1 encompass a wide variety of diseases. Most notably, however, SUV39H1 plays a huge role in cancer and inflammatory disease. Since SUV39H1 is part of a family of histone mthyltransferases, there is a potential use as an epigentic control.
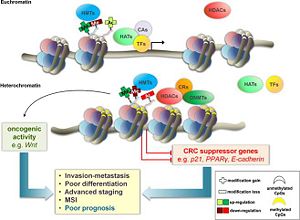 Figure 1 showing the methylation of the histone protein and the subsequent formation of heterochromatin
References
--
|
|
|



