Using Java for Rendering Structures
From Proteopedia
Proteopedia now uses the version of Jmol that supports running with HMTL5/Javascript or the traditional Java. Proteopedia defaults to the HMTL5/Javascript of Jmol.
However, as Java remains the best for performance, when present we encourage you to use Java to render the 3D visualizations on Proteopedia.
Users with Java installed on their computer can select Proteopedia to run Jmol in Java mode on a page-by-page basis or registered users can set Java mode as your default option. How to do each of these two options is explained below.
NOTE: TABLETS AND PORTABLE DEVICES THAT DO NOT HAVE THE OPTION TO INSTALL JAVA ONLY HAVE THE CHOICE OF RUNNING JMOL WITH HMTL5/Javascript. See Java#Detecting your Java version to look into whether you have Java installed.
Rendering Structures with Jmol in Java-mode on a page-by-page basis
Append the code below to the URL of a Proteopedia page to render structures with Jmol using Java.
?_USE=SIGNED
For, example to view the page
http://proteopedia.org/wiki/index.php/G_protein-coupled_receptorusing Java-based Jmol, the URL of the Proteopedia page would be
http://proteopedia.org/wiki/index.php/G_protein-coupled_receptor?_USE=SIGNED
You may now be prompted to accept the certificate for the signed Java applet. Note, you will not see this if you have already selected the checkbox option to remember you trusted it already.
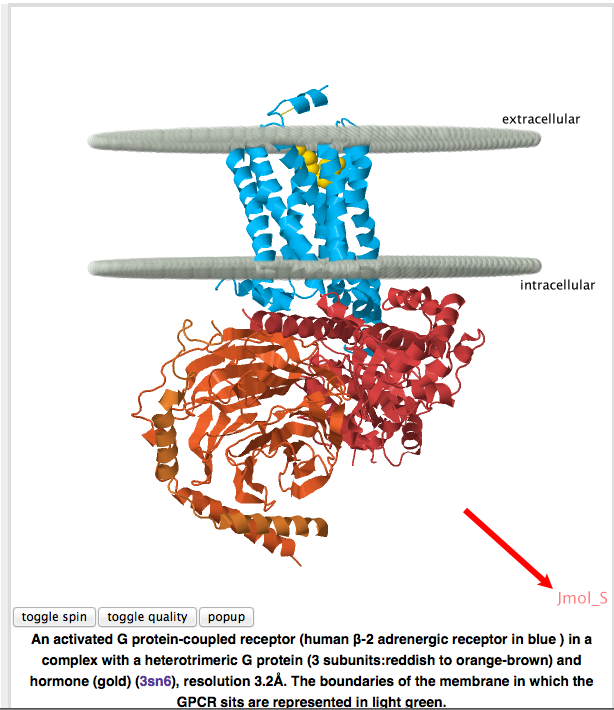
Open a Proteopedia page with a structure on it. When the structure scene appears, you'll see Jmol_S in the bottom right corner of the Proteopedia structure scenes on that page, indicating you are viewing it with the signed Java-based Jmol applet. See here for more information about distinguishing the modes.
Defaulting to rendering Proteopedia's structures with Jmol in Java-mode
Registered users of Protopepedia can set Java mode as your default option for rendering structures on Proteopedia.
How to do this follows:
- Click
Log inin the upper right corner in order to sign in to Proteopedia with your user name and password, if you have not already. If you are not already a registered user, selectLog in/request accountin the upper right corner to request an account.
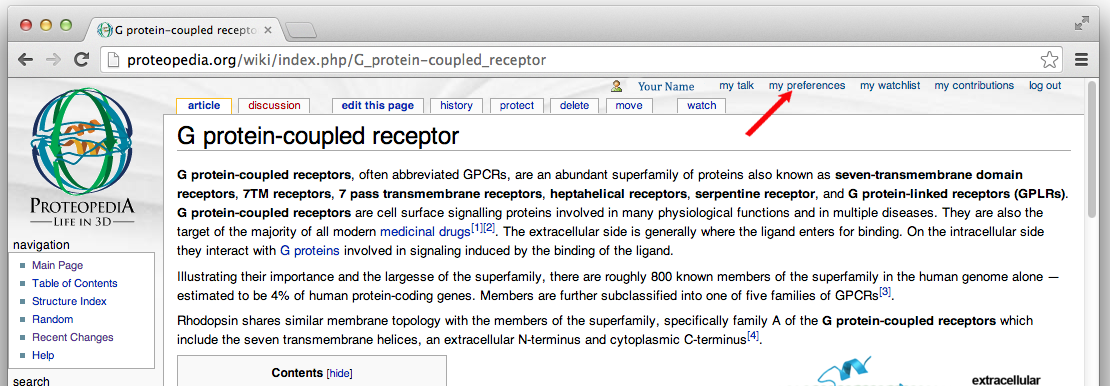
- Select
my preferencesfrom the tool bar in the upper right of each Proteopedia page.
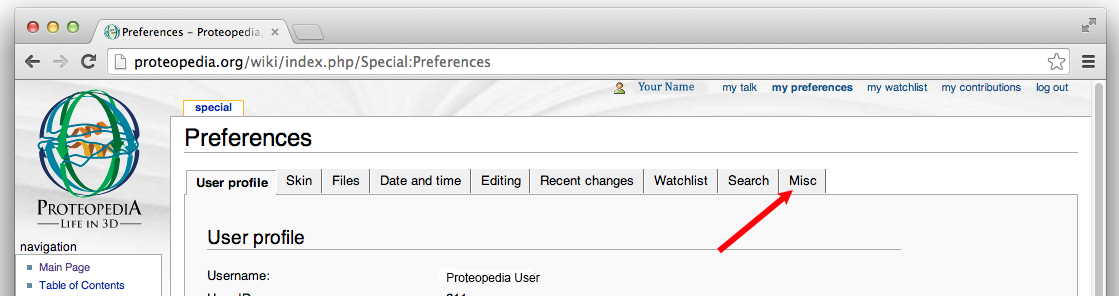
- Select
Misc.from the tabs in the middle of the preference settings page.
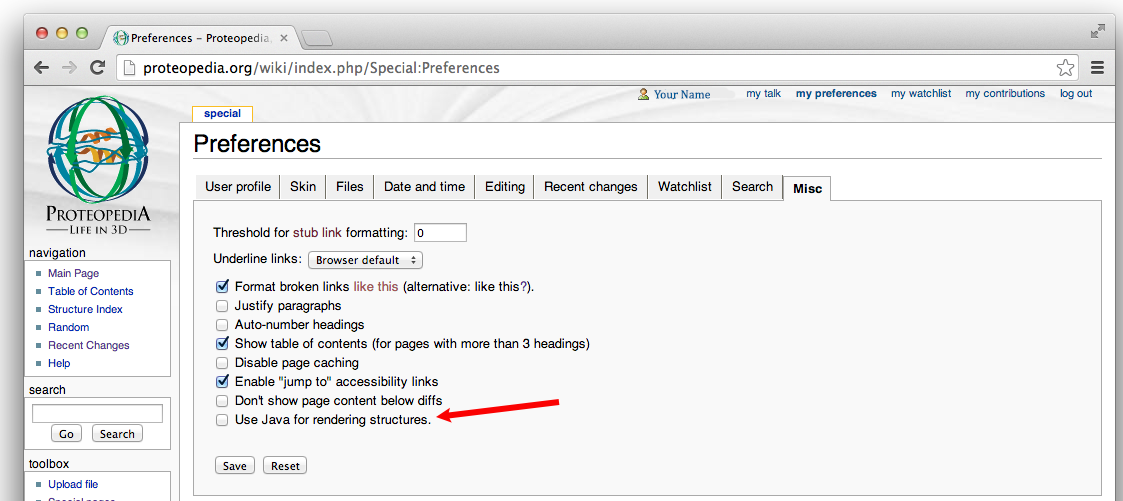
- Look towards the bottom of the options under the
Use Java for rendering structures.
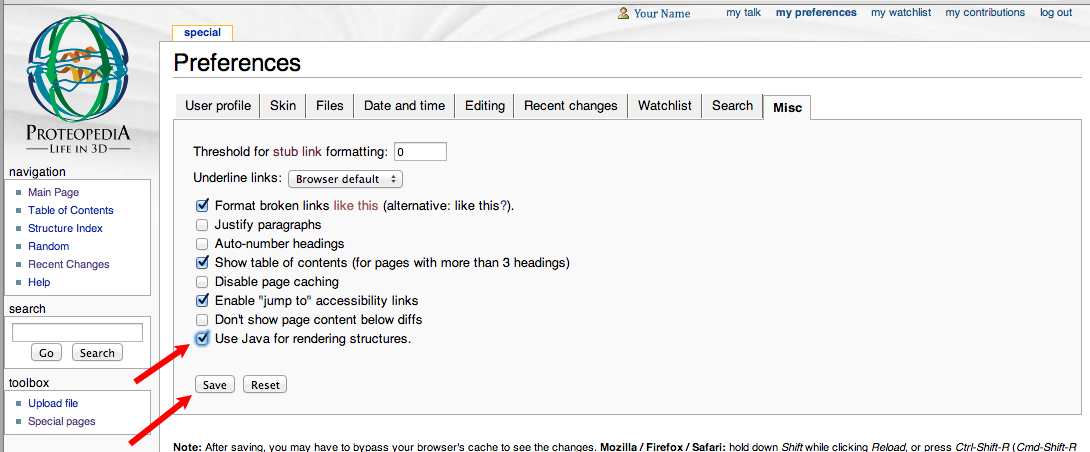
- You want to toggle on the checkbox for
Misc., and then click onSaveto save your new settings.
- Open a Proteopedia page with a structure on it. When the structure scene appears, you'll see Jmol_S in the bottom right corner of the Proteopedia structure scenes on that page, indicating you are viewing it with the signed Java-based Jmol applet.
See here for more information about distinguishing the modes.