This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Image:OB fold.jpg
From Proteopedia
Size of this preview: 800 × 418 pixels
Full resolution (2058 × 1076 pixel, file size: 242 KB, MIME type: image/jpeg)
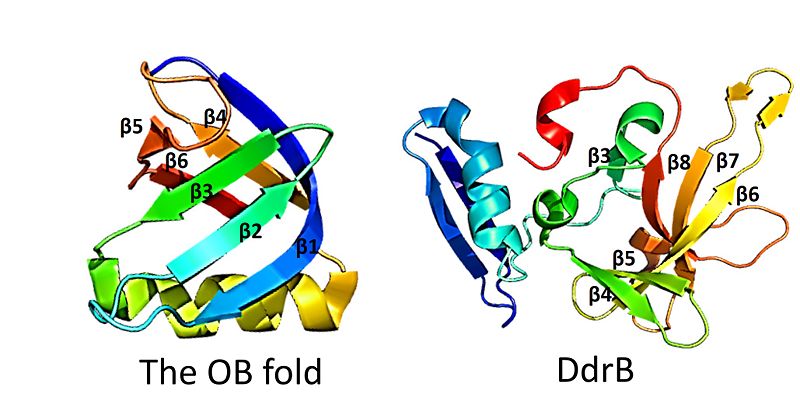
This figure highlights the differences between the classic OB fold found in single-stranded binding proteins and the novel structural features of DdrB. The OB fold is observed in the protein verotoxin-1, PDB code 2XSC . DdrB is modeled from the PDB structure 4HQB. This figure was generated using Pymol.
File history
Click on a date/time to view the file as it appeared at that time.
| Date/Time | User | Dimensions | File size | Comment | |
|---|---|---|---|---|---|
| (current) | 11:29, 30 April 2014 | Lauren Ferris (Talk | contribs) | 2058×1076 | 242 KB | This figure highlights the differences between the classic OB fold found in single-stranded binding proteins and the novel structural features of DdrB. The OB fold is observed in the protein verotoxin-1, PDB code 2XSC . DdrB is modeled from the PDB stru |
- Edit this file using an external application
See the setup instructions for more information.
Links
The following pages link to this file:
