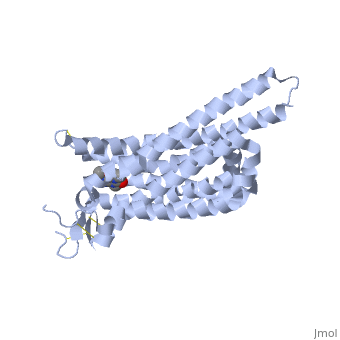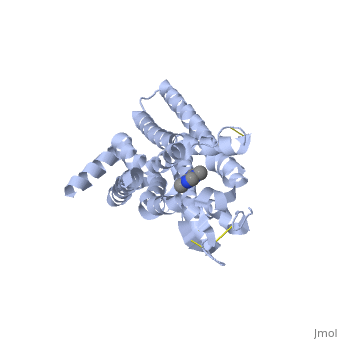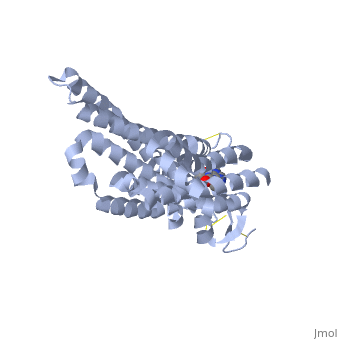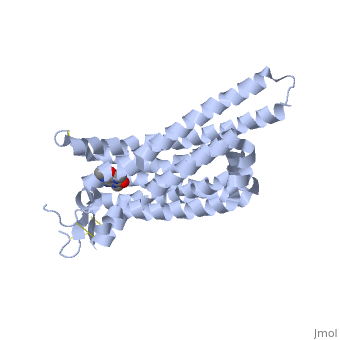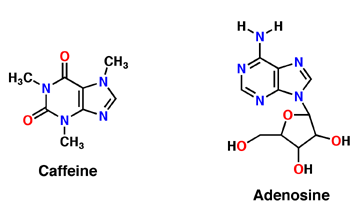
Caffeine (Trimethylxanthine)
Caffeine is a derivative of adenosine and is also called Trimethylxanthine. It is composed of purines; structurally it is polar, and water soluble. They antagonize or inhibit many of the adenosine receptors, like the A1 receptor mentioned above. Caffeine affects neurons and glial cells in the brain by binding to the same location that adenosine would bind and then induce a cascade of enzymatic downstream effects.
Adenosine
Adenosine is an inhibitory neurotransmitter, which promotes sleep and inhibits arousal. It has two components; an adenine nucleotide and a ribose sugar. Adenosine is a polar molecule and is water soluble. Within the brain, concentration of this neuromodulator increases every hour. Adenosine binds extracellularly to G-protein and induces multiple effects. The G-protein is composed of 7 alpha helices, which provide its secondary structure, and is a transmembrane protein. As adenosine receptors bind G-protein, neural activity begins to decrease and the person feels fatigued and sleepy. A2A receptor is one of many adenosine G protein-coupled receptors.
Mechanism of Caffeine (Trimethylxanthine) Synthesis
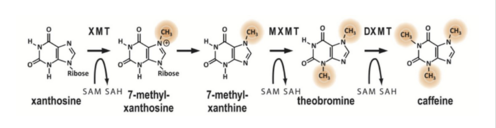
Caffeine is a naturally occurring methylxanthine, purine alkaloid, synthesized by eudicot plants such as coffee, cacao, and tea (Denoeud et. al, 2014). In order to synthesize caffeine, xanthosine must undergo 3 methylation steps with the help of three NMT enzymes; xanthosine methyltransferase (XMT), theobromine synthase (MXMT), and caffeine synthase (DXMT) (Denoeud et. al, 2014). The first step of caffeine biosynthesis involves XMT converting S-adenosylmethionine (SAM) to S-adenosylhomocysteine (SAH) which removes a methyl group and adds it to the 7’-Nitrogen. This produces the intermediate 7-methyl-xanthosine which may undergo resonance to become 7-methyl-xanthine (Denoeud et. al, 2014). The second enzyme, MXMT, converts another SAM to SAH, subsequently add a methyl group to the 3’- Nitrogen on 7-methyl-xanthine. This produces theobromine which may undergo another methylation step with the help of the enzyme DXMT. DXMT converts a third SAM to SAH, adding a methyl group to the 1’-Nitrogen, yielding a caffeine molecule (Denoeud et. al, 2014).
Four Different Adenosine Receptors
There are four different types of adenosine receptors that bind and activate to adenosine, the ligand for that receptor. The four types, A1, A2A, A2B, and A3 are all members of the G protein coupler receptor, which is a membrane spanning protein. These receptors are expressed in the brain, immune system, and cardiovascular system. The receptor, A1, protects the heart from an oxygen deficiency, slowing down the heart rate. When adenosine binds to the A1 receptor it causes a cascade of effects, such as reduces the cyclic AMP level, increase calcium concentration, and increases ERK1 and ERK 2 (kinases that help with cell growth and differentiation) (Antonioli, et. al., 2013).
The A2B receptor is an integral membrane protein that, when in the presence of adenosine, stimulates the presence of adenylate cyclase. A2B also is involved in axon elongation, by interacting with the protein netrin-1 (ADORA2B Adenosine A2b Receptor [ Homo Sapiens (human) ].)The A3 receptor, also a G- protein linked receptor, is involved in the cell growth and division, and cell death. This receptor also has both neurodegenerative and neuroprotective effects ("ADORA3 Adenosine A3 Receptor [ Homo Sapiens (human) ]). The A2A receptor improves the flow of blood to the heart, increasing heart rate, and additionally can lower blood pressure. When adenosine binds to the A2, cyclicAMP levels increase, and ERK1/ERK2 levels increase (Antonioli, et. al., 2013).
Structural highlights
The adenosine receptor (A2A) is a G-protein, which is a transmembrane protein that consists of secondary structures, such as seven alpha helical domains. Inside the third and seventh transmembrane helical domains, there are hydrophobic side chains that are required for ligand recognition. The target ligand, adenosine, is a large, polar molecule that binds to the extracellular binding domain of the A2A receptor by several nonpolar interactions. To be specific, these nonpolar interactions include hydrogen bonding (11), aromatic stacking interactions (1), and many van der Waals interactions (Xu et. al, 2011). To avoid the steric interactions between the ribose ring of adenosine and the tryptophan of the enzyme binding pocket, these nonpolar interactions cause conformational changes within the binding cavity, and cause an internal rotation and tilt of the seventh helical domain (Xu et. al, 2011). Other molecules, such as caffeine can also bind to these adenosine receptors. When caffeine binds to this receptor, it inhibits adenosine from binding to the extracellular binding domain of the A2A receptor.
Caffeine (Trimethylxanthine)
A2A is a transmembrane G protein in humans. Trimethylxanthine has a highly water soluble and thus when present in the system, interacts with the A2A receptor. In order for Trimethylxantine to bind to the receptor, the third and seventh transmembrane helical domains need to recognize the ligand. Trimethylxanthine can then bind. Trimethylxanthine can bind with very little discomfort, due to its similar structure, as well as its purine alkaloid structure, to adenosine. This binding will change the shape and not initiate the cascade of downstream effects that adenosine does, like opening of ion channels and slowing of activity. Concentrate of free adenosine increases extracellularly, when trimethylxanthine is bound. The cAMP increases when adenosine is bound. ERK1 and ERK2 are kinases, composed of serine and threonine, of the GMGC group that regulation of cell growth and differentiation, and if adenosine was bound, this cascade of events would occur, but when Trimethylxanthine is bound, this regulation does not occur.
See Also
http://www.nature.com/nrc/journal/v13/n12/fig_tab/nrc3613_F1.html
http://www.rcsb.org/pdb/explore/explore.do?structureId=4UHR
http://www.rcsb.org/pdb/explore/explore.do?structureId=3RFM
http://www.edb.utexas.edu/ssn/SN%20PDF/Caffeine-Exercise%20Perform.PDF
http://www.ncbi.nlm.nih.gov/pubmed/20164566
http://www.pnas.org/content/112/25/7833.short
http://www.ncbi.nlm.nih.gov/pubmed/1356551