Sandbox WWC8
From Proteopedia
Contents |
Nucleoprotein of Influenza A
|
Introduction
Viruses with anti-sense RNA genomes, such as the influenza virus carry three core polypeptides inside their viral capsids in order to successfully enter a host and initiate the [[viral replication cycle]https://en.wikipedia.org/wiki/Influenza#Replication]. Especially important for viral replication and coordination with the host cell's replication machinery is a protein able to bind single-strand RNA scripts (ssRNA), forming ribonucleoprotein complexes (RNPs). This protein is commonly referred to as nucleoprotein (NP). The NP binds and transports viral RNA scripts to and from the host cell nucleus for transcription, replication, and packaging into new virions. When NP binds RNA, it is structure specific but not sequence specific, meaning NP will bind only ssRNA but will bind any ss-RNA script, viral or non-viral. Beyond the transport function, NP is an essential mediator between host and virus and coordinates complex processes during viral replication [1]. Due to its important function, NP is intensively studies as a potential drug target for antiviral pharmaceuticals [2].
Structural Features
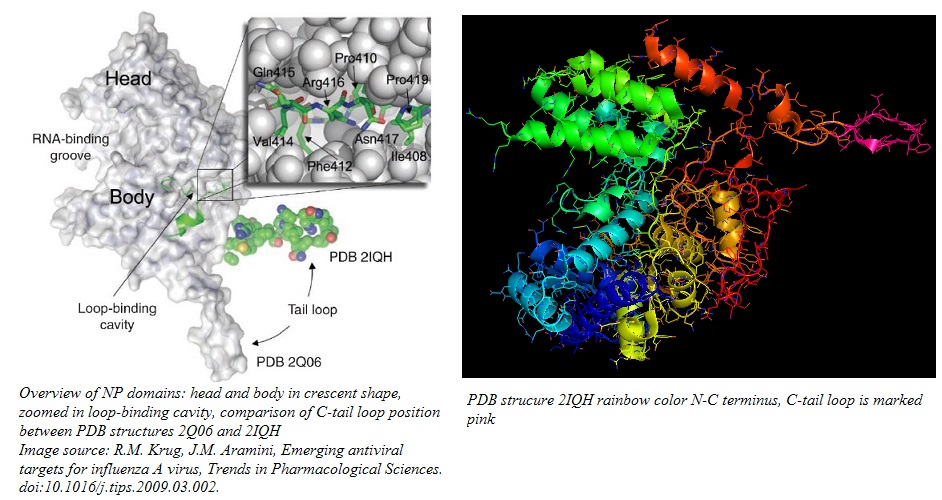
The globular NP protein is rich in arginine, serine, and glycine residues. The abundance of arginine residues gives the protein a net positive charge at pH 7. With a predicted pI of 9.3, NP is mainly composed of basic residues, except for a tail domain of the 30 C-terminal residues, which are acidic. The C-tail domain has a pI of 3.7. The NP is a homo 3-mer - A3 with 499 amino acid residues encoded by the Influenza A RNA segment 5.Each homomer provides a binding site for the viral RNA script. The homomers fold into a crescent shape with a head and a body domain, where the grove between the two domains hosts the ssRNA binding site on the outer surface of the homomer. For the formation of RNPs, the NP needs to oligomerize, forming a coordinated aggregate of three NPs. The C-tail loop, marked in pink in the image below, is critical for this function. Residues 408-419, located at the back of the NP between head and tail domain, form a loop that is the basis of the interface between neighboring NPs. The C-tail loop interlock with the loop binding cavity in the neighboring NP and form a tight binding interaction, featuring both hydrophobic and hydrophilic residues, that keeps the homo 3-mer together. The NP oligomer conformation is especially stabilized by a salt bridge between Arg 416 in the loop and Glu339 in the adjacent NP.
Characterization of Possible Binding Interactions
Role of NP in the Viral Replication cycle
Identification of Antiviral Drug Targets on NP
SHOW INTERACTIVE IMAGE HERE FOR DEFINITION OF RNA BINDING GROOVE RESEARCH WITH NAPROXEN
OTHER POTENTIAL DRUG TARGETS: DISTURBING OLIGOMERIZATION AND NP-POLYMERASE INTERACTION
References
[1] A. Portela, P. Digard, The influenza virus nucleoprotein: a multifunctional RNA-binding protein pivotal to virus replication, Journal of General Virology. 83 (2002) 723–734. doi:10.1099/0022-1317-83-4-723.
[2] R.M. Krug, J.M. Aramini, Emerging antiviral targets for influenza A virus, Trends in Pharmacological Sciences. 30 (2009) 269–277. doi:10.1016/j.tips.2009.03.002.