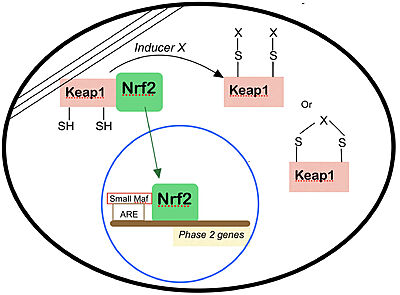
Keap1 is a BTB-Kelch substrate adaptor protein anchored to the actin cytoskeleton that regulates steady-state levels of Nrf2,[1] one of the main transcription factor of cytoprotective genes found in mammals.[2] Under homeostatic conditions, Keap1 is capable of marking Nfr2 for ubiquitin-dependent degradation, repressing the expression of genes associated with cytological defense against highly reactive molecules, such as electrophilic chemicals, heavy metals and oxidative agents. [1] As such, when the cell is subjected to stresses of those kinds, Nrf2 is no longer targeted for ubiquitin-dependent degradation, thus allowing for the expressions of the genes responsible for the cellular defense against such reactive substances.
Relevant Keap1 information summary[3]
| Official Symbol
| KEAP1
|
| Official Full Name
| kelch like ECH associated protein 1
|
| Gene type
| protein coding
|
| RefSeq status
| REVIEWED
|
| Organism
| Homo sapiens
|
| Lineage
| Eukaryota; Metazoa; Chordata; Craniata; Vertebrata; Euteleostomi; Mammalia; Eutheria; Euarchontoglires; Primates; Haplorrhini; Catarrhini; Hominidae; Homo
|
| Expression
| Ubiquitous expression in kidney (RPKM 10.4), prostate (RPKM 9.9) and 25 other tissues (see more)
|
| Orthologs
| Mouse, Cattle, Frog, and more (see more)
|
| Chromosomic Location:
| 19p13.2
|
Function
In eukaryote cells, the defense against reactive molecules from both endogenous or exogenous sources are usually coordinated thanks to the expression of genes regulated by the Cap N' Collar transcription factors, a unique subset within the bZIP family of transcription factors.[4] Such molecules (reactive oxygen species, electrophilic chemicals and heavy metals), if not neutralized and eliminated, can affect the oxidation levels of most endocytic molecules and compromise the integrity of the DNA, leading to severe damage to the organism and causing many pathophysiological processes, including many known diseases in humans such as cancer[5]and cardiovascular problems. [6] The expression of genes regulated by the Cap N' Collar (CNC) set — which includes the transcription factors Nrf1, Nrf2, Nrf3, Bach1 and Bach2 — are indispensable for the induction of enzymes that can neutralize such reactive molecules, eliminate damaged macromolecules and restore cellular redox homeostasis.[1]
substrate adaptor protein responsible for regulating the levels of Nrf2, which together with Nrf1 constitutes the two most prevalent CNC transcription factors in regards to endocytic protection against reactive species in mammals, according to a 2003 experiment that consisted on the observation of the impact of individual CNC genes deletion.[7] Under basal conditions, Keap1 targets Nrf2 for ubiquitin-dependent degradation[1] by bridging the Neh2 domain (a conserved N-terminal regulatory domain) of the Nrf2 to a molecule of , a component of Cullin-RING E3 ubiquitin ligases complexes (CRLs). Once the lysine residues within the Neh2 domain of Nrf2 are marked, cyclical association and dissociation of this E3 ubiquitin ligase complex, mediated by the opposing actions of CAND1 and Cul3 neddylation, enables efficient ubiquitination of Nrf2[1], repressing the expression of Nrf2 dependent genes, reducing the ability of the cell to neutralize reactive molecules. But when the cell is under oxidative stress, a array of are modified,[8] perturbing the ubiquitin ligase complex and razing the levels of Nrf2, thus allowing for the expression of the genes responsible for the cytological defense against the invasive molecule.
Nrf2 is anchored in the cytoplasm by binding to Keap1, which is attached to the actin cytoskeleton[8]. Inducers disrupt the Keap1–Nrf2 complex, and Nrf2 migrates to the nucleus where it forms heterodimers with other transcription factors such as small Maf that bind to the ARE regions of phase 2 genes and accelerate their transcription. Several types of modifications of Keap1 by inducers are shown.
Mechanism and function of Nrf2
Nrf2 belongs to the CNC (Cap ‘n’ Collar) family of b-Zip transcription factors, acting through the formation of a heterodimer with one of the small Maf proteins to activate the expression genes responsible for the defense against xenobiotic and oxidative stresses. [9] Like all CNC family members, Nrf2 is likely to interact with antioxidant responsive elements, manly: genes related to phase II enzymes production, genes related to the xc− system [10] (such as the xCT gene, responsible for encoding the cysteine–glutamate-exchange transporter, which mediates cysteine influx coupled with the efflux of intracellular glutamate) and the gene responsible for Mrp1 production (a ATP-binding cassette transporter that has an important role in the cellular exclusion of conjugated phase II metabolites[9]). Hence, Nrf2 coordinately regulates the xenobiotic conjugation reaction, the supply of intracellular glutathione and the excretion of xenobiotics, enabling efficient detoxification and cytoprotection against xenobiotic toxicity, and studies have shown that mice impaired with Nrf2 deficiency are more susceptible to xenobiotic stress. [11] [12] [13] [14]
According to Motohashi and Yamamoto,[9] Nrf2 is also an important regulator of oxidative-stress inducible genes, including heme oxygenase-1 and peroxiredoxin 1. In a 2002 study[15], a single-nucleotide polymorphism was detected in the promoter region of the Nrf2 gene of the mouse strain C57BL/6J, which is sensitive to hyperoxic stress. Supporting this, Nrf2-null mutant mice were found to be highly susceptible to hyperoxic lung injury. The impaired defense mechanisms against oxidative stress that are observed in the nrf2-null mutant mice could have resulted from the accumulation of reactive oxygen species (ROS) in the absence of Nrf2. A combination of electron paramagnetic resonance (EPR) and spin-probe kinetic analysis confirmed that there is a substantial decrease in the ability of nrf2-null mutant liver and kidney to eliminate ROS. This impaired elimination of ROS was exacerbated in aging female animals. Consistent with this result, old female Nrf2-deficient mice with an ICR genetic background often developed severe lupus-like autoimmune nephritis. Because ROS have a prominent role in the pathogenesis of nephritis, the accumulation of ROS due to Nrf2 deficiency must have exacerbated the mild glomerular lesions that are inherent to the ICR strain of mice.
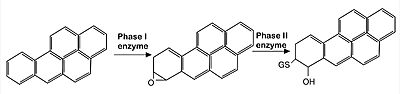
Image: representation of endocytic metabolisation of reactive agents (in this case, Benoz[a]pyrine molecules). The xenobiotic agent is first oxidized by phase I enzymes belonging to the P450 mono-oxygenase system (such as CYP1A1 and CYP1A2 enzymes). Since the product of such reactions are often highly reactive, they are subsequently detoxified by the induction of enzymes related to Phase II reactions (such as glutathione S-transferase and UDP–glucuronosyl-transferase), that promote the conjugation of phase I products with hydrophilic moieties, such as glutathione and glucuronic acid, so that the final, non-reactive product can be then eliminated by excretion. Nrf2 is the main CNC transcription factor responsible for the induction of those Phase II enzymes.
Structural and Genetic information of Nrf2
NRF2 possesses six highly conserved domains called NRF2-ECH homology (Neh) domains. Their names and function is as follows:
Neh1 domain: A CNC-bZIP domain that allows Nrf2 to heterodimerize with small Maf proteins.
: allows for binding of NRF2 to Keap1.
Neh3, Neh4 and Neh5 domain: may play a role in NRF2 protein stability and may act as a transactivation domain, interacting with component of the transcriptional apparatus (Neh3) or to a protein called cAMP Response Element Binding Protein (Neh4 and Neh5), which possesses intrinsic histone acetyltransferase activity.
Neh6 domain: may contain a degron that is involved in a redox-insensitive process of degradation of NRF2. This occurs even in stressed cells, which normally extend the half-life of NRF2 protein relative to unstressed conditions by suppressing other degradation pathways.
The gene responsible for Nrf2 coding in humans is called HAGRID 283, and its sequence of bases is available in the Human Ageing Genomic Resources database, and it is as follows:
Relevance
Structural highlights
Keap1 finalmente todos os dominios:
Nrf2 todos os dominios: