BASIL2023GVQ8DN35
From Proteopedia
Q8DN35 Protein
This is a default text for your page BASIL2023GVQ8DN35. Click above on edit this page to modify. Be careful with the < and > signs. You may include any references to papers as in: the use of JSmol in Proteopedia [1] or to the article describing Jmol [2] to the rescue.
|
Contents |
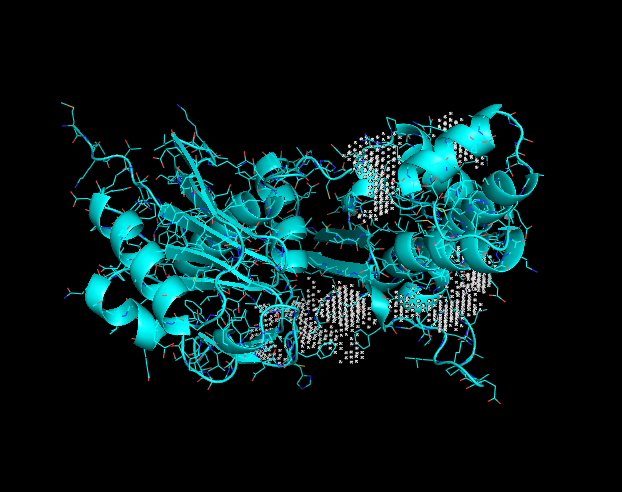
Crystal structure
Function
Through computational docking results, the predicted function for the Q8DN35 protein is that of a glucokinase. The use of tools such as NetGO, BLAST, DALI, InterPRO, PyRX and SPRITE allowed for this predicted function. Q8DN35's sequence is related to that of the ROK Kinase family. NetGO predicted that this protein will have glucokinase activity as it's deepest result. PyRX docking data showed us that NAG (2-acetamido-2-deoxy-beta-D-glucopyranose) and F6P (fructose-6-phosphate) had high binding affinity in the potential binding pocket of Q8DN35.
Binding site
This image displays potential binding sites to the Q8DN35 protein.
Protein purification analysis
Through the use of a gravity purification column and Ni-NTA resin, Q8DN35 was purified and a Bradford analysis was conducted to determine the amount of protein in the purified sample. This purification resulted in 0.055 mg/mL of protein being purified.
Kinase activity analysis
To observe Q8DN35's activity, a kinase activity was ran with 2 substrates to observe potential substrate usage. The 2 substrates that were tested are NAG and F6P.
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644