From Proteopedia
proteopedia linkproteopedia link
| This Sandbox is Reserved from March 18 through September 1, 2025 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson and Mark Macbeth at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1828 through Sandbox Reserved 1846.
|
To get started:
- Click the edit this page tab at the top. Save the page after each step, then edit it again.
- show the Scene authoring tools, create a molecular scene, and save it. Copy the green link into the page.
- Add a description of your scene. Use the buttons above the wikitext box for bold, italics, links, headlines, etc.
More help: Help:Editing
|
Structure
Introduction
What are Minibinders?
Minibinders are small proteins that bind to the spike protein that is involved in the viral infection pathway for SARS-CoV-2. These mini proteins target the interaction between SARS-CoV-2 spike protein and ACE2 receptor as an effective therapeutic strategy, targeting the endocytic pathway [1]. The demand for SARS-CoV-2 therapeutics is high, and the promise these minibinders have shown is substantial. These minibinders were able to reduce the viral burden of SARS-CoV-2 in mice [2]. These proteins were de novo (from scratch) designs to mimic the ACE2 helix, but have a lower dissociation constant (greater affinity for spike protein)[3].
COVID-19 Disease Pathway
Understanding the pathway in which COVID-19 infects the host cell is essential to understanding the mechanism that the COVID-19 spike miniproteins prevent viral entry. The spike protein on the virus' surface will bind to the ACE2 receptor on the surface of the host cell, initiating the endocytosis of the virus into the host cell [4]. Following this step, the virus will ultimately translate viral proteins, initiating an immune response, unless there was a protein to interrupt this pathway [2].
COVID-19 Viral Infection Interruption
The minibinders play a role in inhbiting the viral infection of SARS-CoV-2. ACE2 is a cell-membrane surface receptor that will bind to the spike protein on the surface of the viral protein [4]. The minibinders are able to inhibit this step of the viral infection, inhibiting the endocytosis of the virus into the host cell [1].
Design
These minibinders, LCB1 and AHB2, were designed from "scratch" (de novo) with the intention to mimic the binding of ACE2 to spike protein. Using Rotamer Interaction Field (RIF) Docking, the proteins were able to make the most efficient bonding using the ACE2 and spike protein binding interface [1]. Using Site Saturation Mutagenesis (SSM), every residue in the minibinder's helix scaffold design will be substituted with each of the 20 amino acids, one at a time [5]. Forming SSM libraries, each of the libraries converged on a small number of closely related sequences and from these libraries, the design was selected for LCB1 and AHB2, finding a high affinity for the spike protein's receptor binding domain (RBD) [1].
| This is a default text for your page '. Click above on edit this page' to modify. Be careful with the < and > signs.
You may include any references to papers as in: the use of JSmol in Proteopedia [6] or to the article describing Jmol [7] to the rescue.
This is an image of LCB1 bound to the RBD of a spike protein. [1].
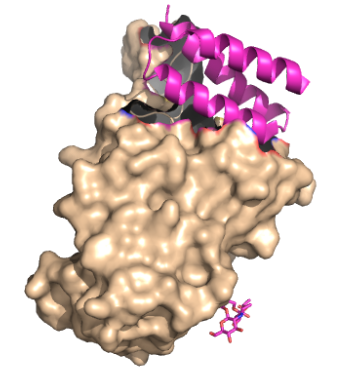 Figure 1. The coolest image ever! This is the link to the salt bridge
Function
Disease
Relevance
Structural highlights
This is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.
|

