We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
FirstGlance/How to get average pLDDT from AlphaFold models
From Proteopedia
It is easy to get the average confidence (pLDDT) for any range of residues in an AlphaFold model by using FirstGlance in Jmol. (Note that AlphaFold Database assigns the same pLDDT value to all atoms in an amino acid, while AlphaFold3 assigns different pLDDT values to each atom in an amino acid.) The procedure here will also report the average estimated error in Å for subsets of residues in models predicted by RoseTTAFold.
- Download the predicted structure as a .pdb file (or .cif file if .pdb is not available). (For the example below, residues 126-761 of human ubiquitin protein ligase E3A Q05086 were submitted to AlphaFold3. See 8jrp at case studies.)
- If only the .cif file is available, convert it to .pdb format.
- Go to FirstGlance in Jmol, and Upload the .pdb file.
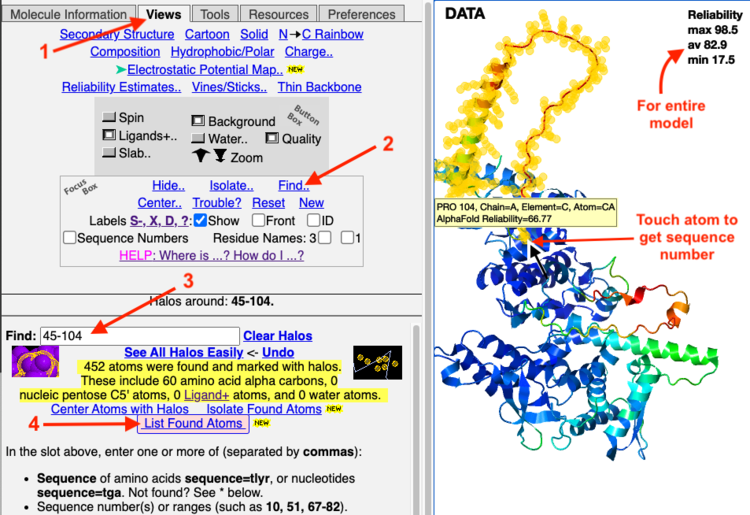
- After the model is displayed in FirstGlance, in the Views tab, click Find.. in the Focus Box:
- Enter the range of sequence numbers in the Find.. slot. (Or enter multiple ranges separated by commas, for example 45-365, 380-682.)
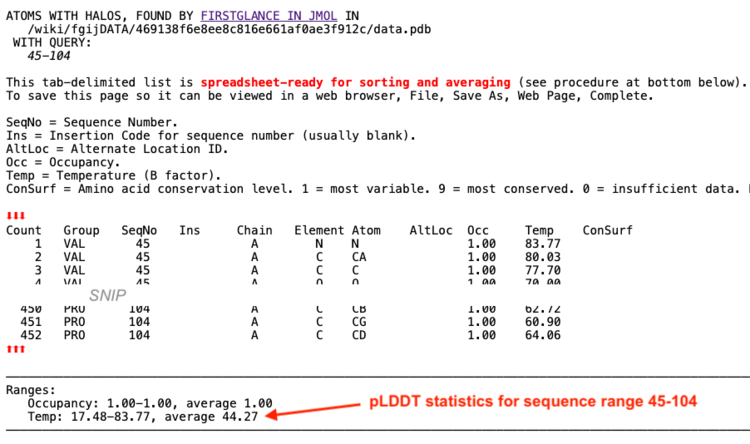
- Click List Found Atoms.
- AlphaFold puts its predicted confidence (pLDDT) in the temperature column, labeled Conf in the listing. The Confidence/pLDDT statistics are at the bottom of the list:
See Also
- AlphaFold/Index, a list of Proteopedia pages about AlphaFold.
- How to predict structures with AlphaFold