FirstGlance in Jmol Literature Citations
From Proteopedia
The following publications in the scientific literature cite use of FirstGlance in Jmol. This list excludes publications co-authored by User:Eric Martz, the author of FirstGlance in Jmol. (Those are listed at Google Scholar: Eric Martz.) The 120 citations listed below in more than 30 peer-reviewed journals were found by searching full text using Google Scholar (see Coverage), and all were verified to cite FirstGlance in Jmol.
|
FirstGlance in Jmol offers, since 2006, free visualization and analysis of protein molecules and other macromolecules (DNA, RNA, oligosaccharides, etc.). It is easy to use and provides built-in guidance and help. In 2024, it was used on average 265 times/day. For more, see
|
Researchers took advantage of unique capabilities of FirstGlance:
- Visual analysis of structures predicted by AlphaFold2, colored by reliability estimates.
- Calculation of average pLDDT for structures predicted by AlphaFold3.
- Seeing locations of missing residues, which affect shape, distribution of polar/hydrophobic surfaces, and may result in missing salt bridges and cation-pi interactions.
- Locating mutations in the 3D model.
- Positions of proteins in lipid bilayer membranes (models from OPM U. Mich.).
- Coloring amino acids by evolutionary conservation (determined by ConSurf) to identify functional sites.
- Rejection of models with Rfree below average for their resolutions.
- Applying sequence numbering to the 3D view.
- Seeing the distribution of hydrophobic vs. polar amino acids.
- Percentages of secondary structure elements.
- Locating residues or regions of interest, using Find to apply yellow halos.
- Distribution and counts of charged residues.
- Visual isolation of domains or regions of interest.
- Identification of contact residues.
Use your browser's Find in page (Control-f / Command-f) to locate studies involving the bold terms above. Some of these capabilities are illustrated with animations generated by FirstGlance at tinyurl.com/movingmolecules.
2021-2025
Education & Communication
- Mokos D, Daniel B. Interactive 3D Objects Enhance Scientific Communication of Structural Data. Chembiochem. 2025 Mar 15;26(6):e202500036. PMID:39976303 doi:10.1002/cbic.202500036
- Rose A, Sehnal D, Goodsell DS, Autin L. Mesoscale explorer: Visual exploration of large-scale molecular models. Protein Sci. 2024 Oct;33(10):e5177. PMID:39291955 doi:10.1002/pro.5177
- Procko K, Bakheet S, Beckham JT, Franzen MA, Jakubowski H, Novak WRP. Modeling an Enzyme Active Site using Molecular Visualization Freeware. J Vis Exp. 2021 Dec 25;(178). PMID:35001912 doi:10.3791/63170
BioMedical Research
2025
- Paulson AR, O'Callaghan M, Zhang XX, Naren N, Schoof M, Hurst MRH. Two transcription factors and one antisense RNA underlie the thermoregulated insect pathogenicity of Yersinia entomophaga MH96. Gene. 2025 Dec 11;980:149955. PMID:41390121 doi:10.1016/j.gene.2025.149955
- Bailo R, Kumar CMS, Singh A, Lund PA, Bavro VN, Bhatt A. MmpL12 transports lipooligosaccharides and impacts virulence in Mycobacterium marinum. Microbiology (Reading). 2025 Oct;171(10):001618. PMID:41060696 doi:10.1099/mic.0.001618
For AlphaFold3-predicted structures, "Average pLDDT scores were calculated using FirstGlance in Jmol."
- Karmakar S, Selvaraj S, Mondal S, Gayen D, Baig MJ. Multi-omics reveals DEAD-box RNA helicase 20 as key protein enhancing wilt resistance in transgenic chickpea. Plant Cell Rep. 2025 Dec 3;44(12):289. PMID:41331102 doi:10.1007/s00299-025-03675-8
- Liu YY, Zhang TT, Zhao Y, Li NW, Malhi KK, Yao X, Li JL. Maternal antibodies to Bartha-K61 vaccine did not protect newborn piglets from challenge with novel recombinant pseudorabies virus. Vet J. 2025 Dec;314:106463. PMID:41047069 doi:10.1016/j.tvjl.2025.106463
- Noureddine J, Nair AM, Espinosa KA, Fan L, Cheng J, Zhao R. Pre-N and C-terminal extension regions of Arabidopsis HSP90.7 regulate the chaperone activity and ER stress response. J Biol Chem. 2025 Oct 8;301(12):110806. PMID:41072766 doi:10.1016/j.jbc.2025.110806
|
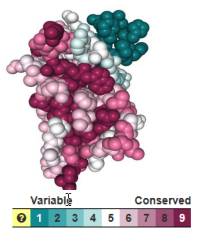
Figure 2 from Sefid et al., 2025. Conservation of amino acids in Teplizumab analyzed using the ConSurf Server and displayed by FirstGlance in Jmol. Reproduced in accord with the Creative Commons Attribution Deed[1] |
|
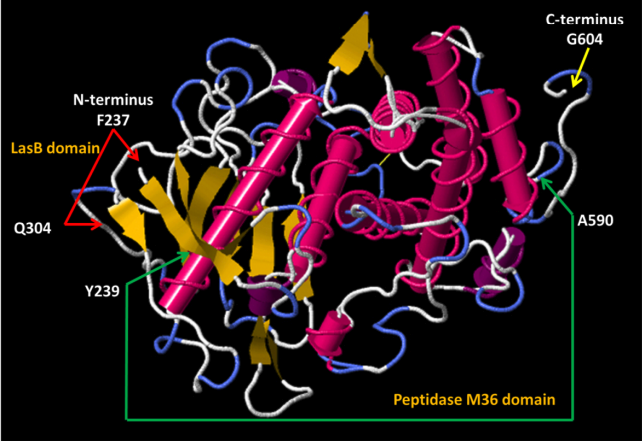
Figure 5 from García, 2025. Molecular rendering of 4x36 by FirstGlance in Jmol. Reproduced with written permission from Ernesto Garcia (April 29, 2025). |
- García E. Structure, Function, and Regulation of LytA: The N-Acetylmuramoyl-l-alanine Amidase Driving the "Suicidal Tendencies" of Streptococcus pneumoniae-A Review. Microorganisms. 2025 Apr 5;13(4):827. PMID:40284663 doi:10.3390/microorganisms13040827
- Gupta M, Kumar H, Debbarma A, Kaur S. Unraveling the abundance of vip3-type genes in Indian Bacillus thuringiensis across the agroclimatic landscape and impact of amino acid substitutions for safer agriculture. Gene. 2025 Jan 15;933:148953. PMID:39299531 doi:10.1016/j.gene.2024.148953
"AlphaFold2 modeling of HIR2 and analysis of the distribution of polar amino acids by FirstGlance revealed an N-terminal surface-exposed platform of HIR2 that consists of nonpolar residues, which could interact with hydrophobic parts of the (plasma) membrane."
- Yao X, Lu WH, Qiao WT, Zhang YQ, Zhang BY, Li HX, Li JL. The highly pathogenic strain of porcine deltacoronavirus disrupts the intestinal barrier and causes diarrhea in newborn piglets. Virulence. 2025 Dec;16(1):2446742. PMID:39758030 doi:10.1080/21505594.2024.2446742
2024
|
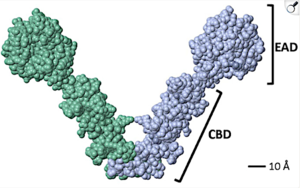
Fig. 1, 1ol5, from Martinez et al., 2024. Molecular rendering and Å scale by FirstGlance in Jmol; black labels added by Martinez et al.. Reproduced in accord with the Creative Commons Attribution Deed[1]. |
- Nabi T, Riyed TH, Ornob A. Deep learning based predictive modeling to screen natural compounds against TNF-alpha for the potential management of rheumatoid arthritis: Virtual screening to comprehensive in silico investigation. PLoS One. 2024 Dec 5;19(12):e0303954. PMID:39636801 doi:10.1371/journal.pone.0303954
FirstGlance in Jmol was used to visualize and tabulate missing residues in 2az5. (See Missing residues and incomplete sidechains.)
- Cerella C, Gajulapalli SR, Lorant A, Gerard D, Muller F, Lee Y, Kim KR, Han BW, Christov C, Récher C, Sarry JE, Dicato M, Diederich M. ATP1A1/BCL2L1 predicts the response of myelomonocytic and monocytic acute myeloid leukemia to cardiac glycosides. Leukemia. 2024 Jan;38(1):67-81. PMID:37904054 doi:10.1038/s41375-023-02076-8
- Rajam SM, Varghese PC, Shirude MB, Syed KM, Devarajan A, Natarajan K, Dutta D. Kinase activity of histone chaperone APLF maintains steady state of centrosomes in mouse embryonic stem cells. Eur J Cell Biol. 2024 Sep;103(3):151439. PMID:38968704 doi:10.1016/j.ejcb.2024.151439
FirstGlance was used to visualize models predicted by AlphaFold2, colored by reliablity scores.
|
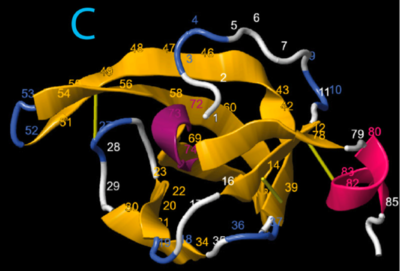
Fig. 3C, and AlphaFold2 prediction, from Gajda et al., 2024. Isolation of this domain, sequence numbering (applied with a single checkbox), and molecular rendering by FirstGlance in Jmol. Reproduced in accord with the Creative Commons Attribution Deed[1]. |
- Gajda Ł, Daszkowska-Golec A, Świątek P. Trophic Position of the White Worm (Enchytraeus albidus) in the Context of Digestive Enzyme Genes Revealed by Transcriptomics Analysis. Int J Mol Sci. 2024 Apr 25;25(9):4685. PMID:38731903 doi:10.3390/ijms25094685
- Li R, Song X, Shan S, Hussain Dhiloo K, Wang S, Yin Z, Lu Z, Khashaveh A, Zhang Y. Female-Biased Odorant Receptor MmedOR48 in the Parasitoid Microplitis mediator Broadly Tunes to Plant Volatiles. J Agric Food Chem. 2024 Aug 7;72(31):17617-17625. PMID:39052973 doi:10.1021/acs.jafc.4c02737
Transmembrane secondary structure segments were evaluated using FirstGlance prior to docking analysis.
FirstGlance was used to locate mutations in the 3D structure.
2023
- García E. Two putative glutamate decarboxylases of Streptococcus pneumoniae as possible antigens for the production of anti-GAD65 antibodies leading to type 1 diabetes mellitus. Int Microbiol. 2023 Aug;26(3):675-690. PMID:37154976 doi:10.1007/s10123-023-00364-y
- Garduño-Sánchez MAA, De Jesus-Bonilla V, Perea S, Miranda-Gamboa R, Herrera-García A, De la Maza Benignos M, Ornelas-García CP. Mitochondrial phylogeography and molecular evolution of the rhodopsin visual pigment in troglobitic populations of Astyanax mexicanus (De Filippi, 1853). Zool Res. 2023 Jul 18;44(4):761-775. PMID:37464933 doi:10.24272/j.issn.2095-8137.2022.437
- Yao X, Qiao WT, Zhang YQ, Lu WH, Wang ZW, Li HX, Li JL. A new PEDV strain CH/HLJJS/2022 can challenge current detection methods and vaccines. Virol J. 2023 Jan 20;20(1):13. PMID:36670408 doi:10.1186/s12985-023-01961-z
2022
|
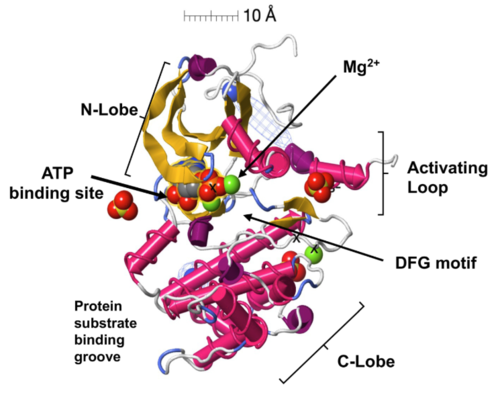
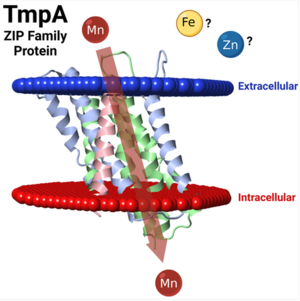
Graphical Abstract from Puccio et al., 2022. Molecular rendering by FirstGlance in Jmol. Red and Blue planes represent boundaries of the lipid bilayer membrane. Reproduced in accord with the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International Deed[2]. |
- Puccio T, Kunka KS, An SS, Kitten T. Contribution of a ZIP-family protein to manganese uptake and infective endocarditis virulence in Streptococcus sanguinis. Mol Microbiol. 2022 Feb;117(2):353-374. PMID:34855265 doi:10.1111/mmi.14853
"Positions within a cellular membrane were predicted using OPM (https://opm.phar.umich.edu/) and visualized in JMol 3.0 using FirstGlance."
- Budania S, Dubey A, Singh A. Trypanosoma evansi RoTat 1.2 variant surface antigen mimotopes selected by panning of the random peptide phage-display library against monoclonal antibodies. J Mol Recognit. 2022 Nov;35(11):e2984. PMID:35869579 doi:10.1002/jmr.2984
- Peter J, Huleux M, Spaniol B, Sommer F, Neunzig J, Schroda M, Li-Beisson Y, Philippar K. Fatty acid export (FAX) proteins contribute to oil production in the green microalga Chlamydomonas reinhardtii. Front Mol Biosci. 2022 Aug 30;9:939834. PMID:36120551 doi:10.3389/fmolb.2022.939834
- Ting TY, Baharin A, Ramzi AB, Ng CL, Goh HH. Neprosin belongs to a new family of glutamic peptidase based on in silico evidence. Plant Physiol Biochem. 2022 Jul 15;183:23-35. PMID:35537348 doi:10.1016/j.plaphy.2022.04.027
|
Figure 5 from Camelo et al., 2025. Molecular rendering by FirstGlance in Jmol. Reproduced in accord with the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International Deed[3]. |
- Quinn JE, Jeninga MD, Limm K, Pareek K, Meißgeier T, Bachmann A, Duffy MF, Petter M. The Putative Bromodomain Protein PfBDP7 of the Human Malaria Parasite Plasmodium Falciparum Cooperates With PfBDP1 in the Silencing of Variant Surface Antigen Expression. Front Cell Dev Biol. 2022 Apr 12;10:816558. PMID:35493110 doi:10.3389/fcell.2022.816558
- Khalilullah H, Agarwal DK, Ahsan MJ, Jadav SS, Mohammed HA, Khan MA, Mohammed SAA, Khan R. Synthesis and Anti-Cancer Activity of New Pyrazolinyl-Indole Derivatives: Pharmacophoric Interactions and Docking Studies for Identifying New EGFR Inhibitors. Int J Mol Sci. 2022 Jun 11;23(12):6548. PMID:35742992 doi:10.3390/ijms23126548
- Al Kufi SGJH, Emmerson J, Rosenqvist H, Garcia CMM, Rios-Szwed DO, Wiese M. Absence of DEATH kinesin is fatal for Leishmania mexicana amastigotes. Sci Rep. 2022 Feb 28;12(1):3266. PMID:35228627 doi:10.1038/s41598-022-07412-z
- Rodríguez-Enríquez A, Herrera-Camacho I, Millán-Pérez-Peña L, Reyes-Leyva J, Santos-López G, Rivera-Benítez JF, Rosas-Murrieta NH. Predicted 3D model of the M protein of Porcine Epidemic Diarrhea Virus and analysis of its immunogenic potential. PLoS One. 2022 Feb 9;17(2):e0263582. PMID:35139120 doi:10.1371/journal.pone.0263582
2021
- Rubin M, Ben-Tal N. Using ConSurf to Detect Functionally Important Regions in RNA. Curr Protoc. 2021 Oct;1(10):e270. PMID:34619810 doi:10.1002/cpz1.270
- Zhang J. Molecular dynamics studies of dog prion protein wild-type and its D159N mutant. J Biomol Struct Dyn. 2021 Aug;39(12):4234-4242. PMID:32496928 doi:10.1080/07391102.2020.1776155
- Loza-Muller L, Shitan N, Yamada Y, Vázquez-Flota F. AmABCB1, an alkaloid transporter from seeds of Argemone mexicana L (Papaveraceae). Planta. 2021 Nov 16;254(6):122. PMID:34786595 doi:10.1007/s00425-021-03780-4
2016-2020
Education & Communication
- Lee NY, Tucker-Kellogg G. An accessible, open-source mobile application for macromolecular visualization using augmented reality. Biochem Mol Biol Educ. 2020 May;48(3):297-303. PMID:32077575 doi:10.1002/bmb.21335
Lee et al., 2020, said "... browser-based applications or interfaces (e.g., FirstGlance in Jmol) increase accessibility for students. Hands-on use of visualization software by students seems to benefit their 3D understanding of proteins better than simply viewing it on-screen."
- Zheng M, Waller MP. ChemPreview: an augmented reality-based molecular interface. J Mol Graph Model. 2017 May;73:18-23. PMID:28214437 doi:10.1016/j.jmgm.2017.01.019
BioMedical Research
2020
|
Fig. 2d from Lin et al., 2020. Secondary structure percentages and molecular rendering by FirstGlance in Jmol. Reproduced in accord with the Creative Commons Attribution-NonCommercial 4.0 International Deed[4]. |
- Lin M, Jin Y, Chen X, Sui Y, Li Y, Li H, Ni X, Zhao N, Lu Y, Jiang M. Increased hydrophobicity of CRYGD p.(Ala159ProfsTer9): Suspected cause of congenital cataracts in a large Chinese family. Mol Genet Genomic Med. 2020 Oct;8(10):e1436. PMID:33460241 doi:10.1002/mgg3.1436
- Ben Chorin A, Masrati G, Kessel A, Narunsky A, Sprinzak J, Lahav S, Ashkenazy H, Ben-Tal N. ConSurf-DB: An accessible repository for the evolutionary conservation patterns of the majority of PDB proteins. Protein Sci. 2020 Jan;29(1):258-267. PMID:31702846 doi:10.1002/pro.3779
Ben Chorin et al., 2020, wrote "The conservation grades (colors) are mapped onto the three-dimensional structure of the query protein, which can be viewed using ... FirstGlance in Jmol. This visualization is highly enlightening because it emphasizes the important, evolutionarily conserved regions of the protein."
- Pollack JD, Gerard D, Makhatadze GI, Pearl DK. Evolutionary conservation and structural localizations suggest a physical trace of metabolism's progressive geochronological emergence. J Biomol Struct Dyn. 2020 Aug;38(12):3700-3719. PMID:31608807 doi:10.1080/07391102.2019.1679666
|
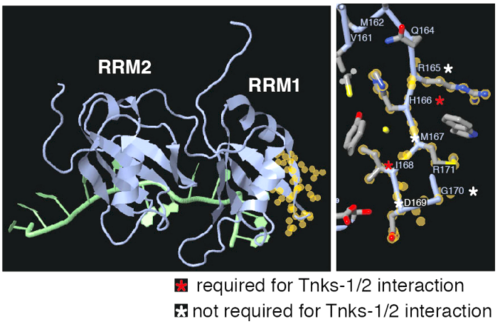
Portions of Fig. 2I,J from McGurk et al., 2020. The tankyrase-binding-domain of 4bs2 was identified (yellow halos) with the Find tool of FirstGlance in Jmol. FirstGlance also added the sequence labels. Reproduced in accord with the Creative Commons Attribution 4.0 International Deed[1]. |
- McGurk L, Rifai OM, Bonini NM. TDP-43, a protein central to amyotrophic lateral sclerosis, is destabilized by tankyrase-1 and -2. J Cell Sci. 2020 Jun 23;133(12):jcs245811. PMID:32409565 doi:10.1242/jcs.245811
- Hong Y, Lee J, Vu TH, Lee S, Lillehoj HS, Hong YH. Immunomodulatory effects of avian β-defensin 5 in chicken macrophage cell line. Res Vet Sci. 2020 Oct;132:81-87. PMID:32531564 doi:10.1016/j.rvsc.2020.06.002
- Shalit Y, Tuvi-Arad I. Side chain flexibility and the symmetry of protein homodimers. PLoS One. 2020 Jul 24;15(7):e0235863. PMID:32706779 doi:10.1371/journal.pone.0235863
Shalit and Tuvi-Arad, 2020, wrote "For each protein [565 were analyzed], we calculated the Rfree grade as defined by FirstGlance in Jmol .... Files were kept if their Rfree grade was at least "average" at their resolution."
- Mattila HK, Mäkinen M, Lundell T. Hypoxia is regulating enzymatic wood decomposition and intracellular carbohydrate metabolism in filamentous white rot fungus. Biotechnol Biofuels. 2020 Feb 24;13:26. PMID:32123543 doi:10.1186/s13068-020-01677-0
Mattila et al. 2020 provided a downloadable PDB file with a link to view it by uploading to FirstGlance in Jmol.
- Hong Y, Lee J, Vu TH, Lee S, Lillehoj HS, Hong YH. Chicken avian β-defensin 8 modulates immune response via the mitogen-activated protein kinase signaling pathways in a chicken macrophage cell line. Poult Sci. 2020 Sep;99(9):4174-4182. PMID:32867961 doi:10.1016/j.psj.2020.05.027
2019
- Eozenou C, Bashamboo A, Bignon-Topalovic J, Merel T, Zwermann O, Lourenco D, Lottmann H, Lichtenauer U, Rojo S, Beuschlein F, McElreavey K, Brauner R. The TALE homeodomain of PBX1 is involved in human primary testis-determination. Hum Mutat. 2019 Aug;40(8):1071-1076. PMID:31058389 doi:10.1002/humu.23780
- Tuvi-Arad I, Alon G. Improved algorithms for quantifying the near symmetry of proteins: complete side chains analysis. J Cheminform. 2019 Jun 6;11(1):39. PMID:31172379 doi:10.1186/s13321-019-0360-9
- Flores BN, Li X, Malik AM, Martinez J, Beg AA, Barmada SJ. An Intramolecular Salt Bridge Linking TDP43 RNA Binding, Protein Stability, and TDP43-Dependent Neurodegeneration. Cell Rep. 2019 Apr 23;27(4):1133-1150.e8. PMID:31018129 doi:10.1016/j.celrep.2019.03.093
- Del Mondo A, Iovinella M, Petriccione M, Nunziata A, Davis SJ, Cioppa D, Ciniglia C. A Spotlight on Rad52 in Cyanidiophytina (Rhodophyta): A Relic in Algal Heritage. Plants (Basel). 2019 Feb 19;8(2):46. PMID:30791384 doi:10.3390/plants8020046
2018
- Wang H, Avnir D, Tuvi-Arad I. Chiral Ramachandran Plots II: General Trends and Protein Chirality Spectra. Biochemistry. 2018 Nov 13;57(45):6395-6403. PMID:30346734 doi:10.1021/acs.biochem.8b00974
- Brand T. The Popeye Domain Containing Genes and Their Function as cAMP Effector Proteins in Striated Muscle. J Cardiovasc Dev Dis. 2018 Mar 13;5(1):18. PMID:29534001 doi:10.3390/jcdd5010018
- Menozzi I, Polverini E, Berni R. Deciphering protein dynamics changes along the pathway of retinol uptake by cellular retinol-binding proteins 1 and 2. Arch Biochem Biophys. 2018 May 1;645:107-116. PMID:29567208 doi:10.1016/j.abb.2018.03.020
- Kaliamurthi S, Selvaraj G, Junaid M, Khan A, Gu K, Wei DQ. Cancer Immunoinformatics: A Promising Era in the Development of Peptide Vaccines for Human Papillomavirus-induced Cervical Cancer. Curr Pharm Des. 2018;24(32):3791-3817. PMID:30398106 doi:10.2174/1381612824666181106094133
- Gallant JP, Lima-Cordón RA, Justi SA, Monroy MC, Viola T, Stevens L. The role of natural selection in shaping genetic variation in a promising Chagas disease drug target: Trypanosoma cruzi trans-sialidase. Infect Genet Evol. 2018 Aug;62:151-159. PMID:29684709 doi:10.1016/j.meegid.2018.04.025
- Liang J, Li L, Jin X, Xu B, Pi L, Liu S, Zhu W, Zhang C, Luan B, Gong L, Zhang C. Pharmacological effect of human melanocortin-2 receptor accessory protein 2 variants on hypothalamic melanocortin receptors. Endocrine. 2018 Jul;61(1):94-104. PMID:29704154 doi:10.1007/s12020-018-1596-2
- Hong Y, Truong AD, Lee J, Lee K, Kim GB, Heo KN, Lillehoj HS, Hong YH. Identification of duck liver-expressed antimicrobial peptide 2 and characterization of its bactericidal activity. Asian-Australas J Anim Sci. 2019 Jul;32(7):1052-1061. PMID:30381731 doi:10.5713/ajas.18.0571
2017
- Naasse Y, Bakhchane A, Charoute H, Jennane F, Bignon-Topalovic J, Malki A, Bashamboo A, Barakat A, Rouba H, McElreavey K. A Novel Homozygous Missense Mutation in the FU-CRD2 Domain of the R-spondin1 Gene Associated with Familial 46,XX DSD. Sex Dev. 2017;11(5-6):269-274. PMID:29262419 doi:10.1159/000485393
- Lu Y, Ondo Y, Shimojima K, Osaka H, Yamamoto T. A novel TUBB4A mutation G96R identified in a patient with hypomyelinating leukodystrophy onset beyond adolescence. Hum Genome Var. 2017 Aug 3;4:17035. PMID:28791129 doi:10.1038/hgv.2017.35
- Legati A, Reyes A, Ceccatelli Berti C, Stehling O, Marchet S, Lamperti C, Ferrari A, Robinson AJ, Mühlenhoff U, Lill R, Zeviani M, Goffrini P, Ghezzi D. A novel de novo dominant mutation in ISCU associated with mitochondrial myopathy. J Med Genet. 2017 Dec;54(12):815-824. PMID:29079705 doi:10.1136/jmedgenet-2017-104822
- Dutta SR, Gauri SS, Ghosh T, Halder SK, DasMohapatra PK, Mondal KC, Ghosh AK. Elucidation of structural and functional integration of a novel antimicrobial peptide from Antheraea mylitta. Bioorg Med Chem Lett. 2017 Apr 15;27(8):1686-1692. PMID:28302399 doi:10.1016/j.bmcl.2017.03.003
- Baruch-Shpigler Y, Wang H, Tuvi-Arad I, Avnir D. Chiral Ramachandran Plots I: Glycine. Biochemistry. 2017 Oct 24;56(42):5635-5643. PMID:28872841 doi:10.1021/acs.biochem.7b00525
- Larralde O, Petrik J. Phage-displayed peptides that mimic epitopes of hepatitis E virus capsid. Med Microbiol Immunol. 2017 Aug;206(4):301-309. PMID:28434129 doi:10.1007/s00430-017-0507-0
- Wu D. Epithelial protein lost in neoplasm (EPLIN): Beyond a tumor suppressor. Genes Dis. 2017 Apr 1;4(2):100-107. PMID:30258911 doi:10.1016/j.gendis.2017.03.002
2016
- Santos-Júnior CD, Veríssimo A, Costa J. The recombination dynamics of Staphylococcus aureus inferred from spA gene. BMC Microbiol. 2016 Jul 11;16(1):143. PMID:27400707 doi:10.1186/s12866-016-0757-9
- Ravindra KC, Trudel LJ, Wishnok JS, Wogan GN, Tannenbaum SR, Skipper PL. Hydroxyphenylation of Histone Lysines: Post-translational Modification by Quinone Imines. ACS Chem Biol. 2016 May 20;11(5):1230-7. PMID:26866676 doi:10.1021/acschembio.5b00923
See Figure 2 which uses FirstGlance in Jmol to highlight critical lysine residues in nucleosome histones with yellow halos. That Figure is not reproduced here because ACS denies permission to non-profit organizations, unless payment is made.
- Zhang J, Wang F, Chatterjee S. Molecular dynamics studies on the buffalo prion protein. J Biomol Struct Dyn. 2016;34(4):762-77. PMID:26043781 doi:10.1080/07391102.2015.1052849
FirstGlance in Jmol was used to locate and count charged residues.
- Schindler RF, Scotton C, French V, Ferlini A, Brand T. The Popeye Domain Containing Genes and their Function in Striated Muscle. J Cardiovasc Dev Dis. 2016 Jun 15;3(2):22. PMID:27347491 doi:10.3390/jcdd3020022
- Brody MJ, Feng L, Grimes AC, Hacker TA, Olson TM, Kamp TJ, Balijepalli RC, Lee Y. LRRC10 is required to maintain cardiac function in response to pressure overload. Am J Physiol Heart Circ Physiol. 2016 Jan 15;310(2):H269-78. PMID:26608339 doi:10.1152/ajpheart.00717.2014
2011-2015
Coverage
|
Coverage is nearly complete for the most recent decade 2016-2025 (above), as far as publications found with the methods used[5] in Google Scholar. For years before 2016 (below), only an arbitrary subset of citations is listed. |
Education & Communication
- Barber NC, Stark LA. Engaging with molecular form to understand function. CBE Life Sci Educ. 2014 Spring;13(1):21-4. PMID:24591499 doi:10.1187/cbe.13-12-0247
Barber & Stark wrote "FirstGlance in Jmol is a simple platform with sophisticated functionality for viewing a molecule's structure with different diagrams, cross-sections, and emphasis on various molecular features."
- Berry C, Board J. A Protein in the palm of your hand through augmented reality. Biochem Mol Biol Educ. 2014 Sep-Oct;42(5):446-9. PMID:24979189 doi:10.1002/bmb.20805
- Craig PA, Michel LV, Bateman RC. A survey of educational uses of molecular visualization freeware. Biochem Mol Biol Educ. 2013 May-Jun;41(3):193-205. PMID:23649886 doi:10.1002/bmb.20693
- Forest KT. Enterotoxigenic Escherichia coli CS1 pilus: not one structure but several. J Bacteriol. 2013 Apr;195(7):1357-9. PMID:23354749 doi:10.1128/JB.00053-13
Forest writes "Structures should be made to come alive in articles. ... [A] straightforward option is to include a link to the NSF-supported Java-based application Firstglance in Jmol that will allow the reader to call up the pdb file in a simple Web-based viewer that is browser independent."
- Jaswal SS, O'Hara PB, Williamson PL, Springer AL. Teaching structure: student use of software tools for understanding macromolecular structure in an undergraduate biochemistry course. Biochem Mol Biol Educ. 2013 Sep-Oct;41(5):351-9. doi: 10.1002/bmb.20718. Epub, 2013 Sep 10. PMID:24019219 doi:http://dx.doi.org/10.1002/bmb.20718
Figure 1 is an excellent example of how a student used FirstGlance in Jmol to visualize hydrophobic cores. That Figure is not reproduced here because obtaining permission from Wiley Publications is complicated and may require payment even for a non-profit educational organization with open access on the Internet.
- Harle M, Towns MH. Students' understanding of external representations of the potassium ion channel protein part II: structure-function relationships and fragmented knowledge. Biochem Mol Biol Educ. 2012 Nov-Dec;40(6):357-63. PMID:23166023 doi:10.1002/bmb.20620
FirstGlance in Jmol was the sole 3D visualization tool provided to students in this study.
BioMedical Research
- Pannkuk EL, Risch TS, Savary BJ. Isolation and identification of an extracellular subtilisin-like serine protease secreted by the bat pathogen Pseudogymnoascus destructans. PLoS One. 2015 Mar 18;10(3):e0120508. PMID:25785714 doi:10.1371/journal.pone.0120508
- Kevelam SH, Bierau J, Salvarinova R, Agrawal S, Honzik T, Visser D, Weiss MM, Salomons GS, Abbink TE, Waisfisz Q, van der Knaap MS. Recessive ITPA mutations cause an early infantile encephalopathy. Ann Neurol. 2015 Oct;78(4):649-58. PMID:26224535 doi:10.1002/ana.24496
|
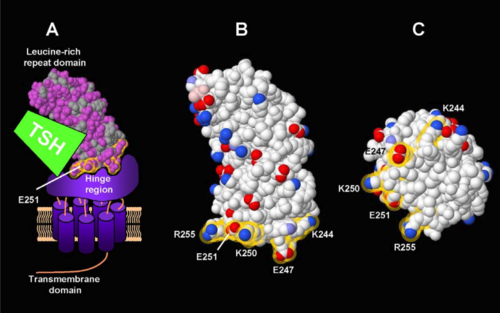
Fig. 1 from Chen et al., 2012. Atomic rendering of the leucine-rich repeat domain of the thyroid-stimulating hormone receptor by FirstGlance in Jmol showing hydrophobic vs. polar regions and charge distribution. Yellow halos highlight charged residues at the C-terminal "base" of the domain. Reproduced in accord with the Creative Commons Attribution 4.0 International Deed[1]. |
- Touw WG, Vriend G. BDB: databank of PDB files with consistent B-factors. Protein Eng Des Sel. 2014 Nov;27(11):457-62. PMID:25336027 doi:10.1093/protein/gzu044
- Barker CS, Meshcheryakova IV, Sasaki T, Roy MC, Sinha PK, Yagi T, Samatey FA. Randomly selected suppressor mutations in genes for NADH:quinone oxidoreductase-1, which rescue motility of a Salmonella ubiquinone-biosynthesis mutant strain. Microbiology. 2014 Apr 1. doi: 10.1099/mic.0.075945-0. PMID:24692644 doi:http://dx.doi.org/10.1099/mic.0.075945-0
- Chen CR, Salazar LM, McLachlan SM, Rapoport B. Novel information on the epitope of an inverse agonist monoclonal antibody provides insight into the structure of the TSH receptor. PLoS One. 2012;7(2):e31973. PMID:22359649 doi:10.1371/journal.pone.0031973
- Lin HE, Tsai WY, Liu IJ, Li PC, Liao MY, Tsai JJ, Wu YC, Lai CY, Lu CH, Huang JH, Chang GJ, Wu HC, Wang WK. Analysis of epitopes on dengue virus envelope protein recognized by monoclonal antibodies and polyclonal human sera by a high throughput assay. PLoS Negl Trop Dis. 2012 Jan;6(1):e1447. PMID:22235356 doi:10.1371/journal.pntd.0001447
- Gao D, Jimenez-Lopez JC, Iwata A, Gill N, Jackson SA. Functional and structural divergence of an unusual LTR retrotransposon family in plants. PLoS One. 2012;7(10):e48595. PMID:23119066 doi:10.1371/journal.pone.0048595
- Taman A, Ribeiro P. Characterization of a truncated metabotropic glutamate receptor in a primitive metazoan, the parasitic flatworm Schistosoma mansoni. PLoS One. 2011;6(11):e27119. PMID:22069494 doi:10.1371/journal.pone.0027119
- Gabanyi MJ, Adams PD, Arnold K, Bordoli L, Carter LG, Flippen-Andersen J, Gifford L, Haas J, Kouranov A, McLaughlin WA, Micallef DI, Minor W, Shah R, Schwede T, Tao YP, Westbrook JD, Zimmerman M, Berman HM. The Structural Biology Knowledgebase: a portal to protein structures, sequences, functions, and methods. J Struct Funct Genomics. 2011 Jul;12(2):45-54. PMID:21472436 doi:10.1007/s10969-011-9106-2
In 2011, the Protein Structure Initiative's Structural Biology Knowledgebase offers FirstGlance in Jmol as a structure viewer.
- Gerlier D. Emerging zoonotic viruses: new lessons on receptor and entry mechanisms. Curr Opin Virol. 2011 Jul;1(1):27-34. PMID:22440564 doi:10.1016/j.coviro.2011.05.014
2006-2010
Education & Communication
- Berry C, Baker MD. Inside protein structures: Teaching in three dimensions. Biochem Mol Biol Educ. 2010 Nov;38(6):425-9. PMID:21567875 doi:10.1002/bmb.20434
... applications such as the excellent, FirstGlance in Jmol provide a quick and simple way to view and manipulate structures ....
- Porollo A, Meller J. POLYVIEW-MM: web-based platform for animation and analysis of molecular simulations. Nucleic Acids Res. 2010 Jul;38(Web Server issue):W662-6. PMID:20504857 doi:10.1093/nar/gkq445
Porollo and Meller wrote "Protein–ligand contacts are determined using the respective procedure adopted in Protein Explorer and subsequently in the FirstGlance in Jmol server (FGiJ) that accounts for hydrogen bonds, water and salt bridges, hydrophobic and aromatic ring interaction and different types of metals binding. For the corresponding bond distance definitions, the reader is referred to the FGiJ documentation." This excellent server continues to be available in 2025 as PolyView-3D, and is linked at the Martz website MolviZ.Org.
- Kumar P, Ziegler A, Grahn A, Hee CS, Ziegler A. Leaving the structural ivory tower, assisted by interactive 3D PDF. Trends Biochem Sci. 2010 Aug;35(8):419-22. PMID:20541422 doi:10.1016/j.tibs.2010.03.008
- O'Donoghue SI, Goodsell DS, Frangakis AS, Jossinet F, Laskowski RA, Nilges M, Saibil HR, Schafferhans A, Wade RC, Westhof E, Olson AJ. Visualization of macromolecular structures. Nat Methods. 2010 Mar;7(3 Suppl):S42-55. doi: 10.1038/nmeth.1427. Epub 2010 Mar, 1. PMID:20195256 doi:http://dx.doi.org/10.1038/nmeth.1427
- Furge LL, Stevens-Truss R, Moore DB, Langeland JA. Vertical and horizontal integration of bioinformatics education: A modular, interdisciplinary approach. Biochem Mol Biol Educ. 2009 Jan;37(1):26-36. PMID:21567685 doi:10.1002/bmb.20249
FirstGlance in Jmol was the primary visualization package provided to students in this curriculum.
- Hodis E, Sussman JL. An encyclopedic effort to make 3D structures easier to understand. Trends Biochem Sci. 2009 Mar;34(3):100-1. Epub 2009 Feb 18. PMID:19230677 doi:10.1016/j.tibs.2009.01.001
Hodis and Sussman wrote "... widely available molecular visualization programs ... are often inaccessible to non-specialists owing to a steep learning curve (with, in our opinion, FirstGlance in Jmol being an exception)."
- Lee WH, Atienza-Herrero J, Abagyan R, Marsden BD. SGC--structural biology and human health: a new approach to publishing structural biology results. PLoS One. 2009 Oct 20;4(10):e7675. PMID:19847312 doi:10.1371/journal.pone.0007675
- McMahon B, Hanson RM. A toolkit for publishing enhanced figures. J Appl Crystallogr. 2008 Aug 1;41(Pt 4):811-814. Epub 2008 Jul 1. PMID:19461848 doi:10.1107/S0021889808015616
Interactivity allows a reader unbounded scope to explore a structure, taking advantage of whatever features the visualization software may provide. This is the basis underlying FirstGlance in Jmol, a service that an increasing number of journals link to, which provides standard buttons to view different aspects of a protein structure ....
- Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
Herráez said "I express my gratitude to Prof. Eric Martz (University of Massachusetts), long-time advocate of molecular modeling teaching, provider of Rasmol and Chime information and support on his website, of teaching-oriented recommendations and sample materials, of the excellent Protein Explorer software for macromolecule visualization and analysis, and of other countless contributions, including the recent FirstGlance in Jmol."
BioMedical Research
- Kotchoni SO, Jimenez-Lopez JC, Gao D, Edwards V, Gachomo EW, Margam VM, Seufferheld MJ. Modeling-dependent protein characterization of the rice aldehyde dehydrogenase (ALDH) superfamily reveals distinct functional and structural features. PLoS One. 2010 Jul 12;5(7):e11516. PMID:20634950 doi:10.1371/journal.pone.0011516
FirstGlance in Jmol was used to visualize patterns of evolutionary conservation calculated by the ConSurf Server.
- Pesi R, Allegrini S, Careddu MG, Filoni DN, Camici M, Tozzi MG. Active and regulatory sites of cytosolic 5'-nucleotidase. FEBS J. 2010 Dec;277(23):4863-72. PMID:21029378 doi:10.1111/j.1742-4658.2010.07891.x
- Goldenberg O, Erez E, Nimrod G, Ben-Tal N. The ConSurf-DB: pre-calculated evolutionary conservation profiles of protein structures. Nucleic Acids Res. 2009 Jan;37(Database issue):D323-7. Epub 2008 Oct 29. PMID:18971256 doi:http://dx.doi.org/10.1093/nar/gkn822
- May M, Brown DR. Diversifying and stabilizing selection of sialidase and N-acetylneuraminate catabolism in Mycoplasma synoviae. J Bacteriol. 2009 Jun;191(11):3588-93. PMID:19329630 doi:10.1128/JB.00142-09
- Nimrod G, Szilágyi A, Leslie C, Ben-Tal N. Identification of DNA-binding proteins using structural, electrostatic and evolutionary features. J Mol Biol. 2009 Apr 10;387(4):1040-53. PMID:19233205 doi:10.1016/j.jmb.2009.02.023
"FirstGlance in Jmol: This free program, also developed by Eric Martz, is probably the easiest way to look at macromolecules."
- Polverini E, Rangaraj G, Libich DS, Boggs JM, Harauz G. Binding of the proline-rich segment of myelin basic protein to SH3 domains: spectroscopic, microarray, and modeling studies of ligand conformation and effects of posttranslational modifications. Biochemistry. 2008 Jan 8;47(1):267-82. PMID:18067320 doi:10.1021/bi701336n
"The "contact residues" of the SH3 domain with the ligand were assigned by means of the FirstGlance in Jmol program ...."
- Nimrod G, Schushan M, Steinberg DM, Ben-Tal N. Detection of functionally important regions in "hypothetical proteins" of known structure. Structure. 2008 Dec 10;16(12):1755-63. PMID:19081051 doi:10.1016/j.str.2008.10.017
- Noto JM, Cornelissen CN. Identification of TbpA residues required for transferrin-iron utilization by Neisseria gonorrhoeae. Infect Immun. 2008 May;76(5):1960-9. PMID:18347046 doi:10.1128/IAI.00020-08
Journals Cited
Peer-reviewed scientific papers listed above that cite FirstGlance in Jmol were published in numerous journals, including
Education Journals
Biochemistry and Molecular Biology Education (8), Journal of Chemical Education.
Basic Research Journals
ACS Chemical Biology, Archives of Biophysics & Biochemistry, Biochemistry (3), BMC Microbiology, Cell Reports, Current Opinion in Virology, European Journal of Cell Biology, FEBS Journal, Gene, Journal of Cell Science, Journal of Bacteriology (2), Journal of Biological Chemistry, Journal of Molecular Biology, Molecular Microbiology, Nature Methods, Nucleic Acids Research (2), Planta, Plant Growth Regulation, Plant Cell Reports, PLOS One (9), Protein Science (3), Structure*, Scientific Reports, Trends in Biochemical Sciences (2), Veterinary Journal.
* Nimrod et al., 2008.
Medical Journals
Annals of Neurology, Infection & Immunity, Leukemia, Medical Microbiology & Immunology, PLOS Neglected Tropical Diseases.
References
- ↑ 1.0 1.1 1.2 1.3 1.4 This work was published under the Creative Commons Attribution 4.0 International Deed.
- ↑ This work was published under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International Deed.
- ↑ This work was published under the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International Deed.
- ↑ This work was published under the Creative Commons Attribution-NonCommercial 4.0 International Deed.
- ↑ In scholar.google.com, the query firstglance finds mostly irrelevant papers with the term first glance. Quoting the query, "firstglance" was used to restrict hits to that exact single word. However, some papers erroneously cite First Glance in Jmol, or even say something like Jmol, with the First Glance. Therefore, a second search was done using "first glance" and jmol.