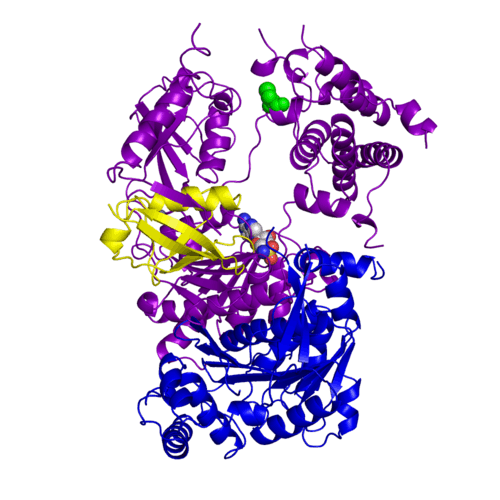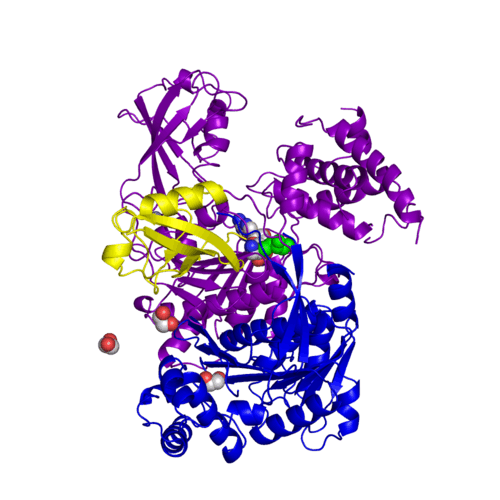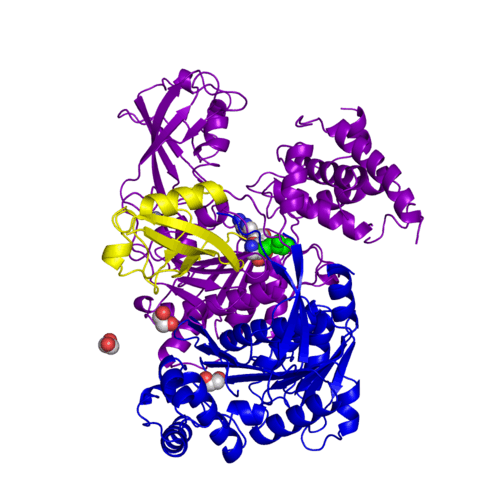
Human SUMO E1 complex with a SUMO1-AMP mimic (3kyc)
Publication Abstract from PubMed
E1 enzymes activate ubiquitin (Ub) and ubiquitin-like (Ubl) proteins in two steps by carboxy-terminal adenylation and thioester bond formation to a conserved catalytic cysteine in the E1 Cys domain. The structural basis for these intermediates remains unknown. Here we report crystal structures for human SUMO E1 in complex with SUMO adenylate and tetrahedral intermediate analogues at 2.45 and 2.6 A, respectively. These structures show that side chain contacts to ATP.Mg are released after adenylation to facilitate a 130 degree rotation of the Cys domain during thioester bond formation that is accompanied by remodelling of key structural elements including the helix that contains the E1 catalytic cysteine, the crossover and re-entry loops, and refolding of two helices that are required for adenylation. These changes displace side chains required for adenylation with side chains required for thioester bond formation. Mutational and biochemical analyses indicate these mechanisms are conserved in other E1s.
Active site remodelling accompanies thioester bond formation in the SUMO E1., Olsen SK, Capili AD, Lu X, Tan DS, Lima CD, Nature. 2010 Feb 18;463(7283):906-12. PMID:20164921
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.
Ubiquitin (Ub) and ubiquitin-like (Ubl) proteins attached to their target proteins and modulating the activities of those targets in various ways. Three types of evolutionarily conserved enzymes — E1 activating enzymes, E2 conjugating enzymes and E3 ligase enzymes — act sequentially through parallel yet distinct pathways to conjugate ubiquitin and Ubl proteins, such as SUMO and NEDD8, to their targets. The E1 enzyme uses the and magnesium to adenylate the C-terminal Ub/Ubl glycine, releasing pyrophosphate and resulting in . A non-hydrolysable and were constructed. In both these compounds the atom of phosphorus is replaced by sulfur (colored yellow).
The of the crystal structures for human SUMO E1 in complex with SUMO adenylate (AMSN) and tetrahedral intermediate (AVSN) analogues revealed opened conformation (SUMO1 in orange, SAE1 colored in blue, and other domains in darkviolet) and closed conformation (SUMO1 in yellow, SAE1 colored in cyan, and other domains in magenta), respectively. In the (3kyc) the distance between Cys domain (including Cys173) and mimic of the acyl adenylate intermediate AMSN is very long, while in the (3kyd), the catalytic Cys173 is posioned near AVSN and SUMO1, so the overall structure revealed dramatic rearrangement. This large conformational change forms the .