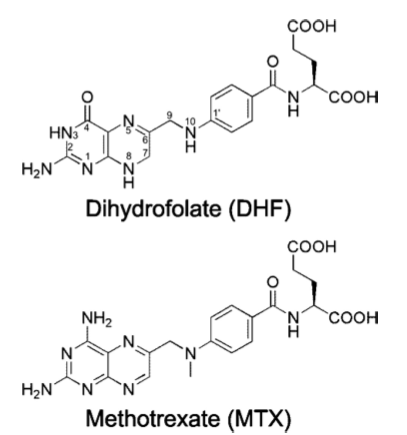
Methotrexate
From Proteopedia
(Difference between revisions)
| Line 49: | Line 49: | ||
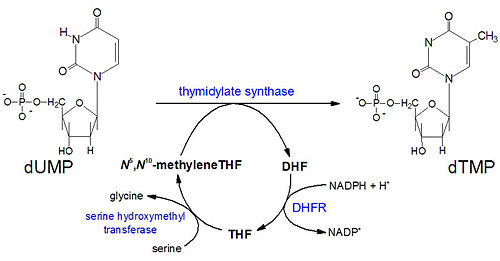
[[Image:DHFR ligands.png|500px|left|thumb| DHFR substrates <ref>Enzymes. (n.d.). Oregon State University. Retrieved March 10, 2011, from http://oregonstate.edu/instruction/bb450/fall2010/lecture/enzymesoutline.html </ref>]] | [[Image:DHFR ligands.png|500px|left|thumb| DHFR substrates <ref>Enzymes. (n.d.). Oregon State University. Retrieved March 10, 2011, from http://oregonstate.edu/instruction/bb450/fall2010/lecture/enzymesoutline.html </ref>]] | ||
| - | + | {{clear}} | |
== Experimental Mutation == | == Experimental Mutation == | ||
| - | |||
| - | <Structure load='3eig' size='500' frame='true' align='right' caption='DHFR methotrexate' scene='Insert optional scene name here' /> | ||
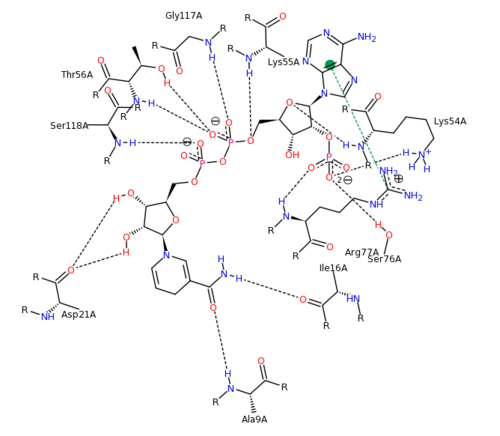
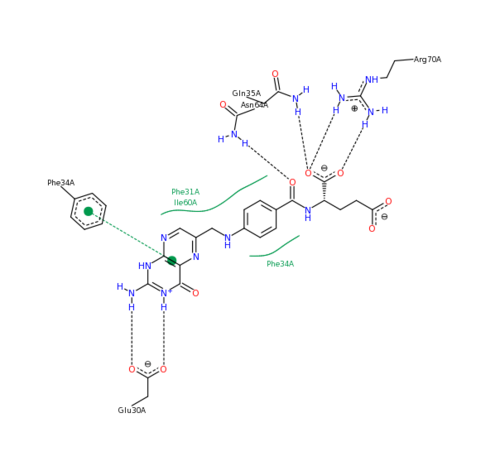
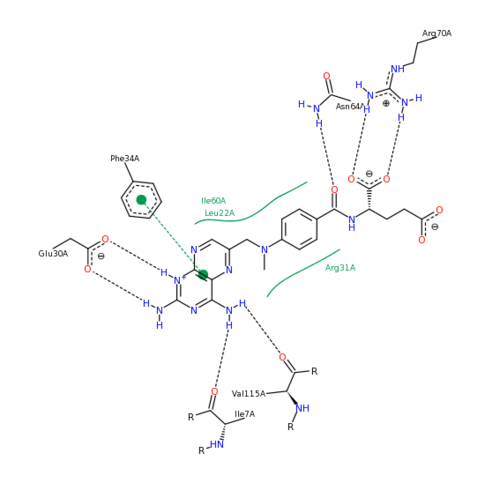
The features of DHFR ligand binding, specifically to methotrexate can be observed and analyzed through various molecular docking and mutation experiments. The a structurally engineered variant of the <scene name='Sandbox_58/Arg_31-35_norm/1'>native human DHFR</scene> altered the F 31 residue of the protein to R, and the Q 35 residue of the protein to E in an attempt to explore the specifics of the methotrexate affinity for DHFR active site residues, resulting in varied active site residues from phenylalanine and glutamine to <scene name='Sandbox_58/Arg_31_glutamine_35/1'>arginine and glutamate</scene>. This mutated enzyme featured a 650x decrease in affinity for the ligand, methotrexate, but retained an amount of methotrexate interaction similar to the enzyme in its native state with native substrates. Crystal structure analysis revealed that the lack of cooperative action and presence of residue disorder lead to the significant decrease in methotrexate activity with the resulting <scene name='Sandbox_58/3eig_active_site/1'>active site</scene>. The arginine residue at place 31, was specifically observed in numerous conformations, a characteristic unique to the mutated enzyme, and the probable cause of the loss of polar contacts and binding affinity between methotrexate and DHFR. A loss of van der Waal forces due to the conformations of the side chains along with an unfavorable placement of Glu-35 causing an “unfavorable electrostatic contact” with methotrexate’s “glutamate portion.” Interestingly this variant was found to display a greater decrease in methotrexate affinity than the decrease in affinity of Dihydrofolate, found to be 9x, evident of catalytic efficiency retention which hold many drug binding resistance implications<ref>Volpato, J., Yachnin, B., & Blanchet, J. (2009). Multiple conformers in active site of human dihydrofolate reductase F31R/Q35E double mutant suggest structural basis for methotrexate resistance.. Journal Biol. Chem., 284, 20079-20089. </ref>. | The features of DHFR ligand binding, specifically to methotrexate can be observed and analyzed through various molecular docking and mutation experiments. The a structurally engineered variant of the <scene name='Sandbox_58/Arg_31-35_norm/1'>native human DHFR</scene> altered the F 31 residue of the protein to R, and the Q 35 residue of the protein to E in an attempt to explore the specifics of the methotrexate affinity for DHFR active site residues, resulting in varied active site residues from phenylalanine and glutamine to <scene name='Sandbox_58/Arg_31_glutamine_35/1'>arginine and glutamate</scene>. This mutated enzyme featured a 650x decrease in affinity for the ligand, methotrexate, but retained an amount of methotrexate interaction similar to the enzyme in its native state with native substrates. Crystal structure analysis revealed that the lack of cooperative action and presence of residue disorder lead to the significant decrease in methotrexate activity with the resulting <scene name='Sandbox_58/3eig_active_site/1'>active site</scene>. The arginine residue at place 31, was specifically observed in numerous conformations, a characteristic unique to the mutated enzyme, and the probable cause of the loss of polar contacts and binding affinity between methotrexate and DHFR. A loss of van der Waal forces due to the conformations of the side chains along with an unfavorable placement of Glu-35 causing an “unfavorable electrostatic contact” with methotrexate’s “glutamate portion.” Interestingly this variant was found to display a greater decrease in methotrexate affinity than the decrease in affinity of Dihydrofolate, found to be 9x, evident of catalytic efficiency retention which hold many drug binding resistance implications<ref>Volpato, J., Yachnin, B., & Blanchet, J. (2009). Multiple conformers in active site of human dihydrofolate reductase F31R/Q35E double mutant suggest structural basis for methotrexate resistance.. Journal Biol. Chem., 284, 20079-20089. </ref>. | ||
| - | [[Image:2011-03-10 2241.png| | + | [[Image:2011-03-10 2241.png|500px|left|thumb| Methotrexate Variant Residue Interaction <ref>DIHYDROFOLATE REDUCTASE COMPLEXED WITH METHOTREXATE. (n.d.). RCSB Protein Database. Retrieved March 10, 2011, from www.rcsb.org/pdb/results </ref>]] |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
== Pharmaceutical Implications == | == Pharmaceutical Implications == | ||
| Line 105: | Line 82: | ||
==Additional Information== | ==Additional Information== | ||
For additional information see: [[Pharmaceutical Drugs]] | For additional information see: [[Pharmaceutical Drugs]] | ||
| - | + | </StructureSection> | |
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 10:16, 23 April 2013
| |||||||||||
References
- ↑ Methotrexate. (n.d.). UW Department of Orthopaedics and Sports Medicine - Patient Care. Retrieved March 10, 2011, from http://www.orthop.washington.edu/PatientCare/OurServices/Arthritis/Articles/Methotrexate.aspx
- ↑ Medical Pharmacology Topics. (n.d.). Angelfire: Welcome to Angelfire. Retrieved March 10, 2011, from http://www.angelfire.com/sc3/toxchick/medpharm/medpharm65.html
- ↑ Methotrexate. (n.d.). UW Department of Orthopaedics and Sports Medicine - Patient Care. Retrieved March 10, 2011, from http://www.orthop.washington.edu/PatientCare/OurServices/Arthritis/Articles/Methotrexate.aspx
- ↑ Schnell JR, Dyson HJ, Wright PE (June 2004). "Structure, dynamics, and catalytic function of dihydrofolate reductase.". Annual Review of Biophysics and Biomolecular Structure 33: 119–40
- ↑ DIHYDROFOLATE REDUCTASE COMPLEXED WITH METHOTREXATE. (n.d.). RCSB Protein Database. Retrieved March 10, 2011, from www.rcsb.org/pdb/results
- ↑ DIHYDROFOLATE REDUCTASE COMPLEXED WITH METHOTREXATE. (n.d.). RCSB Protein Database. Retrieved March 10, 2011, from www.rcsb.org/pdb/results
- ↑ DNA Synthesis - Replication: Chromatin Structure. (n.d.). The Medical Biochemistry Page. Retrieved March 10, 2011, from http://themedicalbiochemistrypage.org/dna.html
- ↑ Voet, D., Voet, J. G., & Pratt, C. W. (2008). Fundamentals of biochemistry: life at the molecular level (3rd ed.). Hoboken, NJ: Wiley.
- ↑ Rajagopalan, P. T. Ravi; Zhang, Zhiquan; McCourt, Lynn (2002). "Interaction of dihydrofolate reductase with methotrexate: Ensemble and single-molecule kinetics". Proceedings of the National Academy of Sciences 99 (21): 13481–6.
- ↑ Methotrexate and Folic Acid. (2006, September 3). Wikimedia Commons. Retrieved March 10, 2011, from commons.wikimedia.org/.png
- ↑ Matthews DA, Alden RA, Bolin JT, Freer ST, Hamlin R, Xuong N, Kraut J, Poe M, Williams M, Hoogsteen K (July 1977). "Dihydrofolate reductase: x-ray structure of the binary complex with methotrexate". Science 197 (4302): 452–455.
- ↑ DIHYDROFOLATE REDUCTASE COMPLEXED WITH METHOTREXATE. (n.d.). RCSB Protein Database. Retrieved March 10, 2011, from www.rcsb.org/pdb/results
- ↑ Enzymes. (n.d.). Oregon State University. Retrieved March 10, 2011, from http://oregonstate.edu/instruction/bb450/fall2010/lecture/enzymesoutline.html
- ↑ Volpato, J., Yachnin, B., & Blanchet, J. (2009). Multiple conformers in active site of human dihydrofolate reductase F31R/Q35E double mutant suggest structural basis for methotrexate resistance.. Journal Biol. Chem., 284, 20079-20089.
- ↑ DIHYDROFOLATE REDUCTASE COMPLEXED WITH METHOTREXATE. (n.d.). RCSB Protein Database. Retrieved March 10, 2011, from www.rcsb.org/pdb/results
- ↑ Methotrexate Information from Drugs.com. (n.d.). Drugs.com | Prescription Drugs - Information, Interactions & Side Effects. Retrieved March 10, 2011, from http://www.drugs.com/methotrexate.html
- ↑ Marks, J. W. (2008, January 8). Methotrexate. Medicine Net. Retrieved March 10, 2011, from www.medicinenet.com/methotrexate/article.htm
- ↑ Trexall. (2007, November 20). The RX List. Retrieved March 10, 2011, from www.rxlist.com/trexall-drug.htm
- ↑ Schwartza, S., & Borner, K. (2007). Glucarpidase (Carboxypeptidase G2) Intervention in Adult and Elderly Cancer Patients with Renal Dysfunction and Delayed Methotrexate Elimination After High-Dose Methotrexate Therapy. The Oncologist, 12(11), 1299-1308.
- ↑ Sirotnak, F., Dorick, D., & Moccio, D. (1978). Murine Tumor ModelsRescue Therapy in the L1210 Leukemia and Sarcoma 180 Optimization of High-Dose Methotrexate with Leucovorin . CANCER RESEARCH, 38, 345-353. Retrieved March 10, 2011, from cancerres.aacrjournals.org/content/38/2/345.full.pdf
- ↑ Methotrexate. (2010, September 1). CCO Formulary. Retrieved March 10, 2011, from www.cancercare.on.ca/pdfdrugs/methotre.pdf
Proteopedia Page Contributors and Editors (what is this?)
Alexander Berchansky, Joel L. Sussman, Daniel Kreider, David Canner, OCA