We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
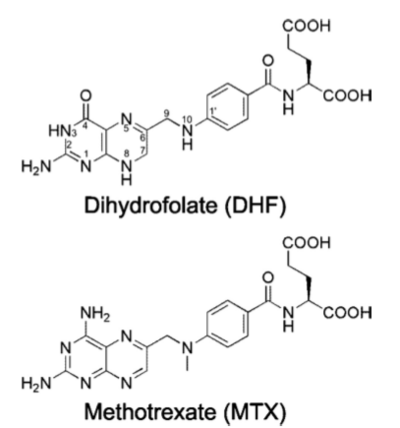
Methotrexate
From Proteopedia
(Difference between revisions)
| (4 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | <StructureSection load='2w3m' size='450' side='right' scene='42/420816/Cv/2' caption=''> | + | <StructureSection load='2w3m' size='450' side='right' scene='42/420816/Cv/2' caption='' pspeed='8’> |
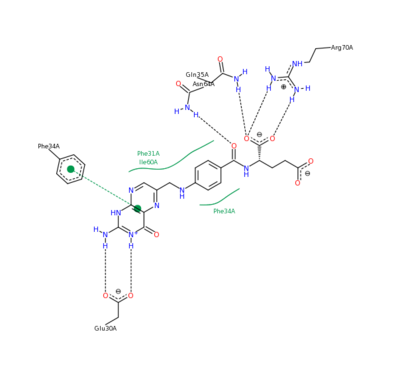
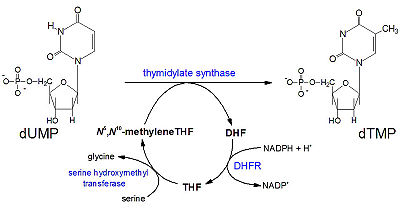
[[Methotrexate]], formerly known as amethopterin, is a drug that is used in the competitive inhibition of [[Dihydrofolate reductase]], resulting in decreased synthesis of dTTP and diminished cellular replication. The antimetabolic nature of methotrexate is most effective against the most rapidly dividing cells, making this drug useful in [[Cancer]] [[Pharmaceutical Drugs|treatment]], and various autoimmune diseases<ref>Methotrexate. (n.d.). UW Department of Orthopaedics and Sports Medicine - Patient Care. Retrieved March 10, 2011, from http://www.orthop.washington.edu/PatientCare/OurServices/Arthritis/Articles/Methotrexate.aspx </ref>. | [[Methotrexate]], formerly known as amethopterin, is a drug that is used in the competitive inhibition of [[Dihydrofolate reductase]], resulting in decreased synthesis of dTTP and diminished cellular replication. The antimetabolic nature of methotrexate is most effective against the most rapidly dividing cells, making this drug useful in [[Cancer]] [[Pharmaceutical Drugs|treatment]], and various autoimmune diseases<ref>Methotrexate. (n.d.). UW Department of Orthopaedics and Sports Medicine - Patient Care. Retrieved March 10, 2011, from http://www.orthop.washington.edu/PatientCare/OurServices/Arthritis/Articles/Methotrexate.aspx </ref>. | ||
__TOC__ | __TOC__ | ||
| Line 44: | Line 44: | ||
| - | The nature of this binding has a 1000 fold increase in affinity relative to the natural folate affinity of DHFR, producing a practically irreversible inhibition of DHFR activity. Methotrexate is a competitive inhibitor that can bind to and inhibit the <scene name='42/420816/Cv/6'>active site of human DHFR</scene> ([[1u72]]). The <scene name='42/420816/Cv/7'>active site is buried within the enzyme</scene>. The <scene name=' | + | The nature of this binding has a 1000 fold increase in affinity relative to the natural folate affinity of DHFR, producing a practically irreversible inhibition of DHFR activity. Methotrexate is a competitive inhibitor that can bind to and inhibit the <scene name='42/420816/Cv/6'>active site of human DHFR</scene> ([[1u72]]). The <scene name='42/420816/Cv/7'>active site is buried within the enzyme</scene>. The <scene name='42/420816/Cv/8'>relative temperature</scene> are color depictions of each atom in regards to mobility or position uncertainty relative to the molecule, with increasing mobility as the color scheme goes from blue to red. |
| Line 52: | Line 52: | ||
== Experimental Mutation == | == Experimental Mutation == | ||
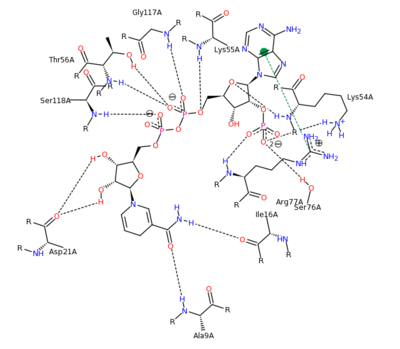
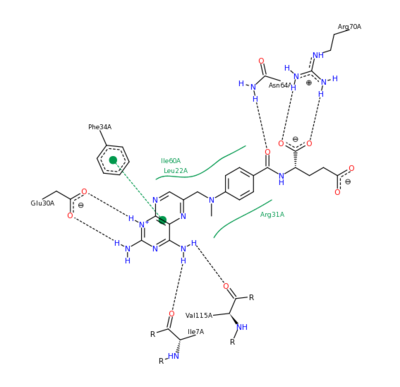
| - | The features of DHFR ligand binding, specifically to methotrexate can be observed and analyzed through various molecular docking and mutation experiments. | + | The features of DHFR ligand binding, specifically to methotrexate can be observed and analyzed through various molecular docking and mutation experiments. The a structurally engineered variant of the <scene name='42/420816/Cv/6'>native human DHFR</scene> ([[1u72]]) altered the <scene name='42/420816/Cv/9'>F 31 residue of the protein to R, and the Q 35 residue of the protein to E</scene> ([[3eig]]) in an attempt to explore the specifics of the methotrexate affinity for DHFR active site residues, resulting in varied active site residues from phenylalanine and glutamine to arginine and glutamate. This mutated enzyme featured a 650x decrease in affinity for the ligand, methotrexate, but retained an amount of methotrexate interaction similar to the enzyme in its native state with native substrates. Crystal structure analysis revealed that the lack of cooperative action and presence of residue disorder lead to the significant decrease in methotrexate activity with the <scene name='42/420816/Cv/10'>resulting active site</scene>. The arginine residue at place 31, was specifically observed in numerous conformations, a characteristic unique to the mutated enzyme, and the probable cause of the loss of polar contacts and binding affinity between methotrexate and DHFR. A loss of van der Waal forces due to the conformations of the side chains along with an unfavorable placement of Glu-35 causing an “unfavorable electrostatic contact” with methotrexate’s “glutamate portion.” Interestingly this variant was found to display a greater decrease in methotrexate affinity than the decrease in affinity of Dihydrofolate, found to be 9x, evident of catalytic efficiency retention which hold many drug binding resistance implications<ref>Volpato, J., Yachnin, B., & Blanchet, J. (2009). Multiple conformers in active site of human dihydrofolate reductase F31R/Q35E double mutant suggest structural basis for methotrexate resistance.. Journal Biol. Chem., 284, 20079-20089. </ref>. |
[[Image:2011-03-10 2241.png|400px|left|thumb| Methotrexate Variant Residue Interaction <ref>DIHYDROFOLATE REDUCTASE COMPLEXED WITH METHOTREXATE. (n.d.). RCSB Protein Database. Retrieved March 10, 2011, from www.rcsb.org/pdb/results </ref>]] | [[Image:2011-03-10 2241.png|400px|left|thumb| Methotrexate Variant Residue Interaction <ref>DIHYDROFOLATE REDUCTASE COMPLEXED WITH METHOTREXATE. (n.d.). RCSB Protein Database. Retrieved March 10, 2011, from www.rcsb.org/pdb/results </ref>]] | ||
| - | + | {{Clear}} | |
== Pharmaceutical Implications == | == Pharmaceutical Implications == | ||
Current revision
| |||||||||||
References
- ↑ Methotrexate. (n.d.). UW Department of Orthopaedics and Sports Medicine - Patient Care. Retrieved March 10, 2011, from http://www.orthop.washington.edu/PatientCare/OurServices/Arthritis/Articles/Methotrexate.aspx
- ↑ Medical Pharmacology Topics. (n.d.). Angelfire: Welcome to Angelfire. Retrieved March 10, 2011, from http://www.angelfire.com/sc3/toxchick/medpharm/medpharm65.html
- ↑ Methotrexate. (n.d.). UW Department of Orthopaedics and Sports Medicine - Patient Care. Retrieved March 10, 2011, from http://www.orthop.washington.edu/PatientCare/OurServices/Arthritis/Articles/Methotrexate.aspx
- ↑ Schnell JR, Dyson HJ, Wright PE (June 2004). "Structure, dynamics, and catalytic function of dihydrofolate reductase.". Annual Review of Biophysics and Biomolecular Structure 33: 119–40
- ↑ DIHYDROFOLATE REDUCTASE COMPLEXED WITH METHOTREXATE. (n.d.). RCSB Protein Database. Retrieved March 10, 2011, from www.rcsb.org/pdb/results
- ↑ DIHYDROFOLATE REDUCTASE COMPLEXED WITH METHOTREXATE. (n.d.). RCSB Protein Database. Retrieved March 10, 2011, from www.rcsb.org/pdb/results
- ↑ DNA Synthesis - Replication: Chromatin Structure. (n.d.). The Medical Biochemistry Page. Retrieved March 10, 2011, from http://themedicalbiochemistrypage.org/dna.html
- ↑ Voet, D., Voet, J. G., & Pratt, C. W. (2008). Fundamentals of biochemistry: life at the molecular level (3rd ed.). Hoboken, NJ: Wiley.
- ↑ Rajagopalan, P. T. Ravi; Zhang, Zhiquan; McCourt, Lynn (2002). "Interaction of dihydrofolate reductase with methotrexate: Ensemble and single-molecule kinetics". Proceedings of the National Academy of Sciences 99 (21): 13481–6.
- ↑ Methotrexate and Folic Acid. (2006, September 3). Wikimedia Commons. Retrieved March 10, 2011, from commons.wikimedia.org/.png
- ↑ Enzymes. (n.d.). Oregon State University. Retrieved March 10, 2011, from http://oregonstate.edu/instruction/bb450/fall2010/lecture/enzymesoutline.html
- ↑ Volpato, J., Yachnin, B., & Blanchet, J. (2009). Multiple conformers in active site of human dihydrofolate reductase F31R/Q35E double mutant suggest structural basis for methotrexate resistance.. Journal Biol. Chem., 284, 20079-20089.
- ↑ DIHYDROFOLATE REDUCTASE COMPLEXED WITH METHOTREXATE. (n.d.). RCSB Protein Database. Retrieved March 10, 2011, from www.rcsb.org/pdb/results
- ↑ Methotrexate Information from Drugs.com. (n.d.). Drugs.com | Prescription Drugs - Information, Interactions & Side Effects. Retrieved March 10, 2011, from http://www.drugs.com/methotrexate.html
- ↑ Marks, J. W. (2008, January 8). Methotrexate. Medicine Net. Retrieved March 10, 2011, from www.medicinenet.com/methotrexate/article.htm
- ↑ Trexall. (2007, November 20). The RX List. Retrieved March 10, 2011, from www.rxlist.com/trexall-drug.htm
- ↑ Schwartza, S., & Borner, K. (2007). Glucarpidase (Carboxypeptidase G2) Intervention in Adult and Elderly Cancer Patients with Renal Dysfunction and Delayed Methotrexate Elimination After High-Dose Methotrexate Therapy. The Oncologist, 12(11), 1299-1308.
- ↑ Sirotnak, F., Dorick, D., & Moccio, D. (1978). Murine Tumor ModelsRescue Therapy in the L1210 Leukemia and Sarcoma 180 Optimization of High-Dose Methotrexate with Leucovorin . CANCER RESEARCH, 38, 345-353. Retrieved March 10, 2011, from cancerres.aacrjournals.org/content/38/2/345.full.pdf
- ↑ Methotrexate. (2010, September 1). CCO Formulary. Retrieved March 10, 2011, from www.cancercare.on.ca/pdfdrugs/methotre.pdf
Proteopedia Page Contributors and Editors (what is this?)
Alexander Berchansky, Joel L. Sussman, Daniel Kreider, David Canner, OCA