Proteopedia:Hot News
From Proteopedia
(Difference between revisions)
| (41 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | + | {| align="right" | |
| - | + | |- | |
| - | + | | | |
| - | [[ | + | <imagemap> |
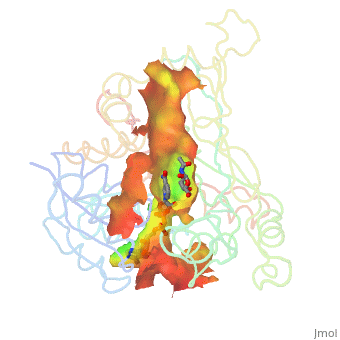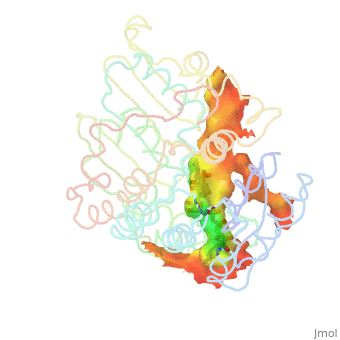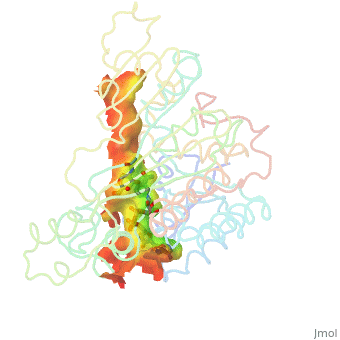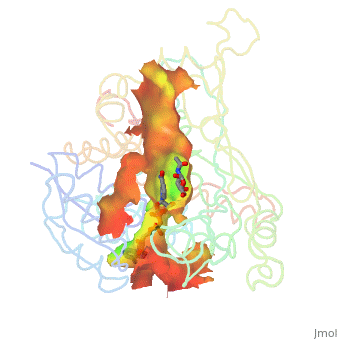
| - | + | Image:Animated_cavity.gif|frame|How to display cavities, pockets, and tunnels. | |
| - | + | default [[Jmol/Cavities_pockets_and_tunnels]] | |
| - | < | + | </imagemap> |
| - | + | |} | |
| - | + | * November 2024: [[How To Find A Structure]] is a new guide to finding an [[empirical models|empirical model]] for a protein of interest, choosing which empirical structure is best, and getting an AlphaFold-predicted structure when no empirical structures are available. | |
| - | + | * November 2024: [[How to predict structures with AlphaFold]] has been updated with instructions for the latest AlphaFold3 Server. | |
| - | < | + | * November 2024: A handful of case studies illustrate what [[Nobel_Prizes_for_3D_Molecular_Structure#2020-2029|Nobel-Prize]] winning AlphaFold3 [[User:Eric_Martz/AlphaFold3_case_studies|can, and cannot, predict]]. |
| - | + | * November 2024: There is a new guide to being aware of, and dealing with [[missing residues and incomplete sidechains]]. | |
| - | + | * March 2024: Professors are using Proteopedia for class projects in Brazil, Czech Republic, France, and various states in the USA. See the updated [[Proteopedia:News#Adoptions_in_College_and_University_Classes|Adoptions in College and University Classes]]. | |
| - | + | * What happens if a SARS-CoV-2 coronavirus enters your lung? See a clear explanation at [[Lifecycle of SARS-CoV-2]] | |
| - | + | * July 2022: <i class="fas fa-walking"></i> <i class="fas fa-running"></i> [[Exercise-induced N-lactoyl-phenylalanine, appetite and obesity]] <i class="fas fa-running"></i> <i class="fas fa-walking"></i> | |
| - | + | * <i class="far fa-star bg-yellow"></i> <b>5,000 users!!</b> <i class="far fa-star bg-yellow"></i> On December 4, 2021, the number of Proteopedia users went over 5,000. The 5,000th user is from Juniata College, Huntingdon, Pennsylvania, US | |
| - | + | * ''AlphaFold protein structure predictions - a step change for biology'', an electronic talk by EBI staff for students and early career researchers via FEBS Junior Sections, Oct. 26, 2021, 19:00 CEST. [https://network.febs.org/posts/alphafold-protein-structure-predictions-a-step-change-for-biology Announcement]. [https://us02web.zoom.us/meeting/register/tZApduCurT0tGNLtFy1Vx4URQ3_kgxLlh2hX Registration]. | |
| - | + | * A practical guide to teaching with Proteopedia <ref>doi 10.1002/bmb.21548</ref> | |
| - | + | * January 2021: How to display cavities, pockets, and tunnels? [[Jmol/Cavities_pockets_and_tunnels]] | |
| - | + | * December 2020: What is changing on SARS-CoV-2 virus and what this means for humanity? [[SARS-CoV-2 spike protein mutations]]. | |
| - | + | * August 2020: An animation of coronavirus spike protein showing the [[SARS-CoV-2 spike protein fusion transformation]]. | |
| - | [[ | + | * July 2020: An animation of coronavirus spike protein showing [[SARS-CoV-2 protein S priming by furin]]. |
| + | * March 2020: New Proteopedia page [http://proteopedia.org/w/Coronavirus_Disease_2019_(COVID-19) focused on the Coronavirus 2019 (COVID-19)]. | ||
| + | * January 2019: New [http://proteopedia.org/cgi-bin/morph morphing] feature for Proteopedia, powered by PyMOL from from Schrodinger. | ||
| + | * January, 2018: [[10th Anniversary Celebration Conference]], University of Massachusetts, Amherst, USA. [[Proteopedia:Scrapbook|Group Photo of Participants]]. | ||
| + | * September, 2017: A workshop based on Proteopedia was held at the [https://www.weizmann.ac.il/conferences/NHBMB2017 New Horizons in Biochemistry & Molecular Biology Education] Conference, jointly organised by FEBS and IUBMB and hosted at the Weizmann Institute of Science. | ||
| + | * June, 2016: [[Help:Making animations for Powerpoint|Animate any Proteopedia scene in Powerpoint]]. | ||
| + | * June, 2016: Proteopedia uses the [[Biological_Unit|Biological Assemblies]] from [http://www.ebi.ac.uk/pdbe PDBe] as the default scenes for all PDB entry pages. Thus, based on the curation by [http://www.ebi.ac.uk EBI] (host for PDBe) the most biologically significant structure is shown. | ||
| + | * June, 2015: [[Proteopedia:Scrapbook#Awards|Award Ceremony]] for the the Proteopedia Award at the ICSG2015 - Deep Sequencing Meets Structural Biology, awarded on 10-Jun-2015 at the Weizmann Institute of Science | ||
| + | * December, 2014, [http://confchem.ccce.divched.org/2014FallCCCENLP9 Talking about Proteopedia] on 12/04/14-12/06/14, a live online talk organised by DivCHED CCCE: Committee on Computers in Chemical Education | ||
| + | * October, 2014 [http://bit.ly/P14UAH Course] in Spanish/English on Proteopedia and its uses to study, display and teach macromolecules. | ||
| + | * [[Create_fast_pages|How to create fast loading pages]] in Proteopedia. | ||
| + | * [[Java#How_to_be_as_safe_as_possible_with_Java|How to be as safe as possible with Java]] | ||
| + | * [[Main_Page_News#Proteopedia_on_iPads!|Proteopedia on iPads!]] | ||
| + | * [[Main_Page_News#What_version_of_Jmol_is_running?|What version of Jmol is running?]] | ||
| + | * [[Main_Page_News#Proteopedia_status|Proteopedia status]] | ||
| + | * [[Main_Page_News#Awards|Awards]] | ||
| + | <references/> | ||
Current revision
- November 2024: How To Find A Structure is a new guide to finding an empirical model for a protein of interest, choosing which empirical structure is best, and getting an AlphaFold-predicted structure when no empirical structures are available.
- November 2024: How to predict structures with AlphaFold has been updated with instructions for the latest AlphaFold3 Server.
- November 2024: A handful of case studies illustrate what Nobel-Prize winning AlphaFold3 can, and cannot, predict.
- November 2024: There is a new guide to being aware of, and dealing with missing residues and incomplete sidechains.
- March 2024: Professors are using Proteopedia for class projects in Brazil, Czech Republic, France, and various states in the USA. See the updated Adoptions in College and University Classes.
- What happens if a SARS-CoV-2 coronavirus enters your lung? See a clear explanation at Lifecycle of SARS-CoV-2
- July 2022: Exercise-induced N-lactoyl-phenylalanine, appetite and obesity
- 5,000 users!! On December 4, 2021, the number of Proteopedia users went over 5,000. The 5,000th user is from Juniata College, Huntingdon, Pennsylvania, US
- AlphaFold protein structure predictions - a step change for biology, an electronic talk by EBI staff for students and early career researchers via FEBS Junior Sections, Oct. 26, 2021, 19:00 CEST. Announcement. Registration.
- A practical guide to teaching with Proteopedia [1]
- January 2021: How to display cavities, pockets, and tunnels? Jmol/Cavities_pockets_and_tunnels
- December 2020: What is changing on SARS-CoV-2 virus and what this means for humanity? SARS-CoV-2 spike protein mutations.
- August 2020: An animation of coronavirus spike protein showing the SARS-CoV-2 spike protein fusion transformation.
- July 2020: An animation of coronavirus spike protein showing SARS-CoV-2 protein S priming by furin.
- March 2020: New Proteopedia page focused on the Coronavirus 2019 (COVID-19).
- January 2019: New morphing feature for Proteopedia, powered by PyMOL from from Schrodinger.
- January, 2018: 10th Anniversary Celebration Conference, University of Massachusetts, Amherst, USA. Group Photo of Participants.
- September, 2017: A workshop based on Proteopedia was held at the New Horizons in Biochemistry & Molecular Biology Education Conference, jointly organised by FEBS and IUBMB and hosted at the Weizmann Institute of Science.
- June, 2016: Animate any Proteopedia scene in Powerpoint.
- June, 2016: Proteopedia uses the Biological Assemblies from PDBe as the default scenes for all PDB entry pages. Thus, based on the curation by EBI (host for PDBe) the most biologically significant structure is shown.
- June, 2015: Award Ceremony for the the Proteopedia Award at the ICSG2015 - Deep Sequencing Meets Structural Biology, awarded on 10-Jun-2015 at the Weizmann Institute of Science
- December, 2014, Talking about Proteopedia on 12/04/14-12/06/14, a live online talk organised by DivCHED CCCE: Committee on Computers in Chemical Education
- October, 2014 Course in Spanish/English on Proteopedia and its uses to study, display and teach macromolecules.
- How to create fast loading pages in Proteopedia.
- How to be as safe as possible with Java
- Proteopedia on iPads!
- What version of Jmol is running?
- Proteopedia status
- Awards
- ↑ Castro C, Johnson RJ, Kieffer B, Means JA, Taylor A, Telford J, Thompson LK, Sussman JL, Prilusky J, Theis K. A practical guide to teaching with Proteopedia. Biochem Mol Biol Educ. 2021 Jun 3. doi: 10.1002/bmb.21548. PMID:34080750 doi:http://dx.doi.org/10.1002/bmb.21548
Proteopedia Page Contributors and Editors (what is this?)
Eric Martz, Jaime Prilusky, David Canner, Joel L. Sussman, Eran Hodis