< User:Eran Hodis(Difference between revisions)
|
|
| (8 intermediate revisions not shown.) |
| Line 1: |
Line 1: |
| - | ===General Structure===
| + | <!--Banner Across Top of Page--> |
| - | There are two distinct classes of HMGRs, class I, which is only found in eukaryotes and are membrane bound and class II, which is found in prokaryotes and are soluble. <ref>PMID:11349148</ref> HMGR contains 8 transmembrane domains, which have yet to be successfully crystallized, that anchor the protein to the membrane of the endoplasmic reticulum. <ref name="Roitelman">PMID:1374417</ref> The catalytic portion of human HMGR forms a tetramer, with the individual monomers winding around each other. <ref name="Roitelman"/> Within the tetramer, the monomers are arranged into <scene name='HMG-CoA_Reductase/1dq8_2_dimers/1'>two dimers</scene>, each of which contains <scene name='HMG-CoA_Reductase/1dq8_2_active_sites/1'>two active sites </scene>which are formed by residues form both monomers. Each monomer contains <scene name='HMG-CoA_Reductase/1dq8_star3_domains/1'>three domains </scene>, the <scene name='HMG-CoA_Reductase/1dq8_n_domain/2'>N-domain</scene>, the <scene name='HMG-CoA_Reductase/1dq8_l_domain/1'>L-Domain</scene>, and the <scene name='HMG-CoA_Reductase/1dq8_s_domain/1'>S-Domain</scene>. The L-domain is unique to HMGRs while the S-domain, which forms the binding site for NADP, resembles that of [[ferredoxin]]. The S and L domains are connected by a <scene name='HMG-CoA_Reductase/1dq8_cis_loop/1'>“cis-loop”</scene> which is essential for the HMG-binding site. <ref name="Roitelman"/> Salt bridges between residues R641 and E782 as well as <scene name='HMG-CoA_Reductase/1dq8_cis_loop/2'>hydrogen bonds</scene> between E700 and E700 on neighboring monomers compliment the largely hydrophobic dimer-dimer interface. <ref name="Roitelman"/>
| + | {| id="Banner" style="width:100%; background:#F5F5FC; margin-top:1.2em; margin-left:0.4em; border:1px solid #ddd;" |
| - | <br />
| + | | style="width:61%; color:#000;" | |
| | | | |
| | + | <!--Welcome Banner--> |
| | + | {| style="width:280px; border:none; background:none;" |
| | + | | style="width:280px; text-align:center; white-space:nowrap; color:#000;" | |
| | + | <div style="font-size:2.0em; border:none; margin:0; padding:0.3em; color:#000;">Welcome to Proteopedia</div> |
| | + | <div style="top:+0.2em; font-size:1.2em; padding-left:5px">The free, collaborative 3D-encyclopedia of proteins & other molecules</div> |
| | + | |} |
| | | | |
| - | ===Substrate Binding & Catalytic Mechanism=== | + | <!-- Links In Upper Banner --> |
| - | [[Image: Reactin_scheme.PNG|300px|left|thumb| Chemical Reaction Catalyzed by HMGR]] | + | | style="width:13%; font-size:0.95em;" | |
| - | The HMG-CoA and NADPH molecules make numerous contacts with the L and S domains in forming the four active sites. The CoA is located in a <scene name='HMG-CoA_Reductase/1dqa_nadp_and_coa/1'>positively charged pocket near the enzyme surface</scene>, with the pantothenic acid moiety extending into the interior of the protein. <scene name='HMG-CoA_Reductase/1dqa_tyr_491/2'>Tyrosine 479 forms a hydrophobic lid</scene> over the CoA adenine base, shielding the extended binding pocket from solution. The NADPH binding site is formed primarily by the S-domain with <scene name='HMG-CoA_Reductase/1dqa_loop/2'>a loop region</scene> playing a critical role in binding. <ref name="Roitelman"/>
| + | * [[Proteopedia:About|About]] |
| | + | * [[Proteopedia:Video_Guide|Video Guide]] |
| | | | |
| - | The HMG binding pocket is the site of catalysis in HMGR. <scene name='HMG-CoA_Reductase/1dqa_cis_loop2/2'> The“cis-loop” that bends over the top of HMG </scene> is a critical structural element of this binding site. Residues <scene name='HMG-CoA_Reductase/1dqa_e_and_d/2'>E559 and D767</scene> and are positioned in the active site as is <scene name='HMG-CoA_Reductase/1dqa_k691/2'>K691 which is only 2.7 angstroms from the HMG O2 carbonyl oxygen</scene>. It is this K691 that presumably stabilizes the negatively charged oxygen on the first mevaldyl-CoA intermediate. <ref name="Roitelman"/> The mevaldyl CoA intermediate is subsequently converted to Mavaldehyde with added stabilization from <scene name='HMG-CoA_Reductase/1dqa_h866/2'>H866, which is within hydrogen bonding distance of the thiol group</scene>. It is then believed that the close proximity of <scene name='HMG-CoA_Reductase/1dqa_e_and_d/2'>E559 and D767</scene> increases the pKA of E559, allowing it to be a proton donor for the reduction of mevaldehyde into mevalonate. <ref name="Roitelman"/>
| + | | style="width:13%; font-size:0.95em;" | |
| - | <br /> | + | * [[Proteopedia:Table of Contents|Table of Contents]] |
| | + | * [[Proteopedia:Structure Index|Structure Index]] |
| | + | |
| | + | | style="width:13%; font-size:0.95em;" | |
| | + | * [[Help:Editing|Editing]] |
| | + | * [[Help:Contents|Help]] |
| | + | |
| | + | |} |
| | + | |
| | + | <!--Left Side: This Weeks Featured Article, and Other Featured Articles--> |
| | + | {| id="Banner 2" style="width:100%; margin:6px; border-spacing: 0px;" |
| | + | | class="Main Left" style="width:65%; border:1px solid #cef2e0; background:#F5FCF5; vertical-align:top; color:#000;" | |
| | + | |
| | + | {| id="Lefts" style="width:100%; vertical-align:top; background:#F5FCF5;" |
| | + | ! style="padding:2px;" | <h2 id="TWFA" style="margin:3px; background:#D9FCDA; font-size:1.25em; font-weight:bold; border:1px solid #a3bfb1; text-align:left; color:#000; padding:0.2em 0.4em;">Featured Article</h2> |
| | + | |- |
| | + | | style="color:#000;padding:2px 5px 5px;" | <div id="Trial"> {{Proteopedia:Featured article/{{#expr: {{#time:U}} mod {{Proteopedia:Number of featured articles}}}}}}</div> |
| | + | |- |
| | + | ! style="padding:2px;" | <h2 id="B" style="margin:3px; background:#D9FCDA; font-size:1.25em; font-weight:bold; border:1px solid #a3bfb1; text-align:left; color:#000; padding:0.2em 0.4em;">Browse Proteopedia</h2> |
| | + | |- |
| | + | | style="color:#000;" | <div id="Browse Proteopedia" style="padding:2px 5px;">{{MainPageBrowse}}</div> |
| | + | |- |
| | + | ! style="padding:2px" | <h2 id="WCPDFU" style="margin:3px; background:#D9FCDA; font-size:1.25em; font-weight:bold; border:1px solid #a3bfb1; text-align:left; color:#000; padding:0.2em 0.4em;">What Can Proteopedia Do For You?</h2> |
| | + | |- |
| | + | | style="color:#000;" | <div id="What Can Proteopedia Do For You" style="padding:2px 5px;">{{MainPageWCPDFU}}</div> |
| | + | |} |
| | + | |
| | + | | style="border:1px solid transparent;" | |
| | + | <!--Right, Proteopedia News and Contributions and Links--> |
| | + | | class="Main Right" style="width:35%; border:1px solid #cedff2; background:#F5F5FC; vertical-align:top;"| |
| | + | |
| | + | {| id="right" style="width:100%; vertical-align:top; background:#F5F5FC;" |
| | + | ! style="padding:2px;" | <h2 id="PN" style="margin:3px; background:#D7D7FE; font-size:1.25em; font-weight:bold; border:1px solid #a3b0bf; text-align:left; color:#000; padding:0.2em 0.4em;">Proteopedia News</h2> |
| | + | |- |
| | + | | style="color:#000;padding:2px 5px;" | <div id="News">{{Template:MainPageNews}}</div> |
| | + | |- |
| | + | ! style="padding:2px;" | <h2 id="Scoreboard" style="margin:3px; background:#D7D7FE; font-size:1.25em; font-weight:bold; border:1px solid #a3b0bf; text-align:left; color:#000; padding:0.2em 0.4em;">The Scoreboard</h2> |
| | + | |- |
| | + | | style="background:#F5F5FC;padding:2px 5px 5px;" | <div id="Contribute"><center>{{Special:ContributionScores}}Score based on pages-edited and number-of-edits,<br>and excludes members of the Proteopedia Team<br>[[Special:ContributionScores|All scores...]]</center></div> |
| | + | |- |
| | + | ! style="padding:2px;" | <h2 id="WTC" style="margin:3px; background:#D7D7FE; font-size:1.25em; font-weight:bold; border:1px solid #a3b0bf; text-align:left; color:#000; padding:0.2em 0.4em;">Want To Contribute?</h2> |
| | + | |- |
| | + | | style="color:#000;padding:2px 5px 5px;" | <div id="Contribute">{{Template:MainPageContribute}}</div> |
| | + | |- |
| | + | ! style="padding:2px;" | <h2 id="PAP" style="margin:3px; background:#D7D7FE; font-size:1.25em; font-weight:bold; border:1px solid #a3b0bf; text-align:left; color:#000; padding:0.2em 0.4em;">Read The Article</h2> |
| | + | |- |
| | + | | style="color:#000;padding:2px 5px 5px;" | <div id="Publications">{{Template:MainPageProteopediaReferences}}</div> |
| | + | |- |
| | + | ! style="padding:2px;" | <h2 id="Contact" style="margin:3px; background:#D7D7FE; font-size:1.25em; font-weight:bold; border:1px solid #a3b0bf; text-align:left; color:#000; padding:0.2em 0.4em;">Contact Us</h2> |
| | + | |- |
| | + | | <div style='font-size:1.2em; text-align:center; vertical-align:center; padding-top:1.0em; padding-bottom:1.0em;'>contact@proteopedia.org</div> |
| | + | |} |
| | + | |} |
| | + | {{Clear}} |
| | + | __NOEDITSECTION__ |
| | + | __NOTOC__ |
Featured Article
|
Green links change the 3D image!
Click and drag on the molecule!
|
|
|
|
|
|
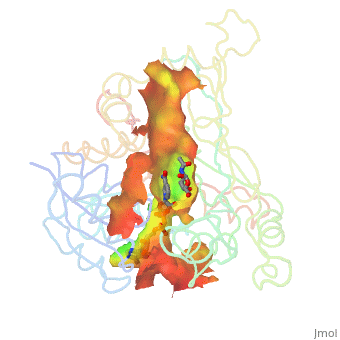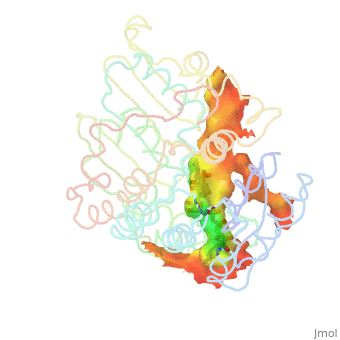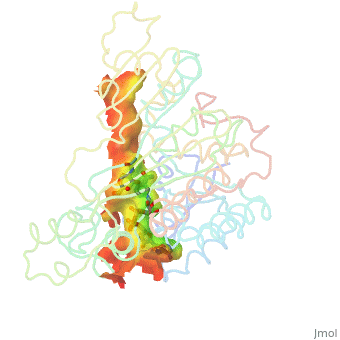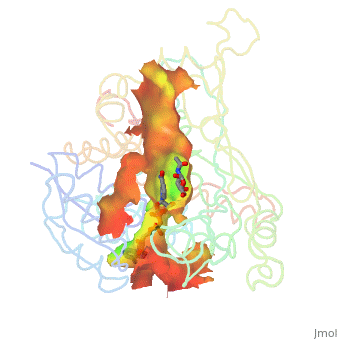
H5N1 bird flu has seemed a likely pandemic threat for decades, but the first new influenza virus to emerge as an imminent pandemic threat in the 21st century is H1N1 swine flu. The drug oseltamivir (Tamiflu®) inhibits flu neuraminidase, a component necessary for virus spread, in susceptible flu strains. Luckily H1N1 swine flu is susceptible (at least in early May, 2009).
The development of oseltamivir was guided, in part, by crystallographically determined structures of flu neuraminidase. Neuraminidase is a homotetramer, shown with oseltamivir bound (). Here is . Oseltamivir was designed to fit N2/N9 (neuraminidases from other strains of flu). Serendipitously, it also fits N1, doing so by (induced fit). The most common mutation in N1 that confers resistance to oseltamivir is H274Y. The mutant tyrosine prevents oseltamivir from fitting, but still allows . (more...)
|
Browse Proteopedia
|
|
Find Your Protein or Biomolecule
|
|
- All PDB entries (over 100,000) have pages
- Go takes you directly to the page if it exists,
- Search gives you search results
|
|
|
What Can Proteopedia Do For You?
|
|
|
|
|
Proteopedia News
|
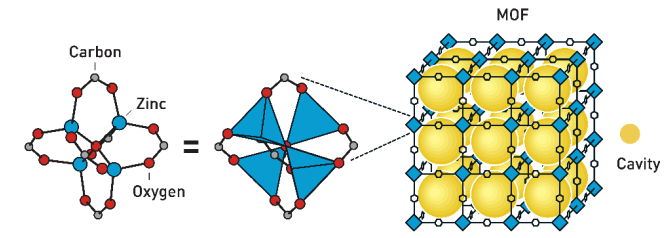
- October 2025: The Nobel Prize in Chemistry recognizes the development of Metal-Organic Frameworks that have a multitude of potential applications. These are similar to Metal-Ligand Polyhedra, but they form extended crystalline networks instead of discrete polyhedra.
- November 2024: How To Find A Structure is a new guide to finding an empirical model for a protein of interest, choosing which empirical structure is best, and getting an AlphaFold-predicted structure when no empirical structures are available.
- November 2024: How to predict structures with AlphaFold has been updated with instructions for the latest AlphaFold3 Server.
- November 2024: A handful of case studies illustrate what Nobel-Prize winning AlphaFold3 can, and cannot, predict.
- November 2024: There is a new guide to being aware of, and dealing with missing residues and incomplete sidechains.
- March 2024: Professors are using Proteopedia for class projects in Brazil, Czech Republic, France, and various states in the USA. See the updated Adoptions in College and University Classes.
- What happens if a SARS-CoV-2 coronavirus enters your lung? See a clear explanation at Lifecycle of SARS-CoV-2
- July 2022: Exercise-induced N-lactoyl-phenylalanine, appetite and obesity
- 5,000 users!! On December 4, 2021, the number of Proteopedia users went over 5,000. The 5,000th user is from Juniata College, Huntingdon, Pennsylvania, US
- AlphaFold protein structure predictions - a step change for biology, an electronic talk by EBI staff for students and early career researchers via FEBS Junior Sections, Oct. 26, 2021, 19:00 CEST. Announcement. Registration.
- A practical guide to teaching with Proteopedia [1]
How to display cavities, pockets, and tunnels. |
- January 2021: How to display cavities, pockets, and tunnels? Jmol/Cavities_pockets_and_tunnels
- December 2020: What is changing on SARS-CoV-2 virus and what this means for humanity? SARS-CoV-2 spike protein mutations.
- August 2020: An animation of coronavirus spike protein showing the SARS-CoV-2 spike protein fusion transformation.
- July 2020: An animation of coronavirus spike protein showing SARS-CoV-2 protein S priming by furin.
- March 2020: New Proteopedia page focused on the Coronavirus 2019 (COVID-19).
- January 2019: New morphing feature for Proteopedia, powered by PyMOL from from Schrodinger.
- January, 2018: 10th Anniversary Celebration Conference, University of Massachusetts, Amherst, USA. Group Photo of Participants.
- September, 2017: A workshop based on Proteopedia was held at the New Horizons in Biochemistry & Molecular Biology Education Conference, jointly organised by FEBS and IUBMB and hosted at the Weizmann Institute of Science.
- June, 2016: Animate any Proteopedia scene in Powerpoint.
- June, 2016: Proteopedia uses the Biological Assemblies from PDBe as the default scenes for all PDB entry pages. Thus, based on the curation by EBI (host for PDBe) the most biologically significant structure is shown.
- June, 2015: Award Ceremony for the the Proteopedia Award at the ICSG2015 - Deep Sequencing Meets Structural Biology, awarded on 10-Jun-2015 at the Weizmann Institute of Science
- December, 2014, Talking about Proteopedia on 12/04/14-12/06/14, a live online talk organised by DivCHED CCCE: Committee on Computers in Chemical Education
- October, 2014 Course in Spanish/English on Proteopedia and its uses to study, display and teach macromolecules.
- How to create fast loading pages in Proteopedia.
- How to be as safe as possible with Java
- Proteopedia on iPads!
- What version of Jmol is running?
- Proteopedia status
- Awards
- ↑ Castro C, Johnson RJ, Kieffer B, Means JA, Taylor A, Telford J, Thompson LK, Sussman JL, Prilusky J, Theis K. A practical guide to teaching with Proteopedia. Biochem Mol Biol Educ. 2021 Jun 3. doi: 10.1002/bmb.21548. PMID:34080750 doi:http://dx.doi.org/10.1002/bmb.21548
|
The Scoreboard
|
Score based on pages-edited and number-of-edits,
and excludes members of the Proteopedia Team
All scores...
|
Want To Contribute?
|
Pages are easy to create and edit,
and green links are fun & easy to make!
Step 1:
Step 2:
Step 3:
|
Read The Article
|
|
|
|
|
| contact@proteopedia.org
|
|