We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
SUMO
From Proteopedia
(Difference between revisions)
| (16 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
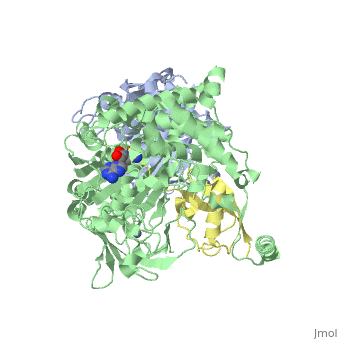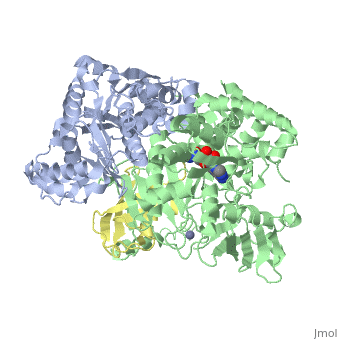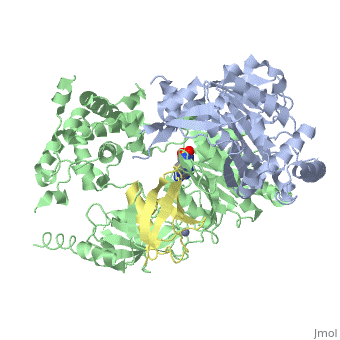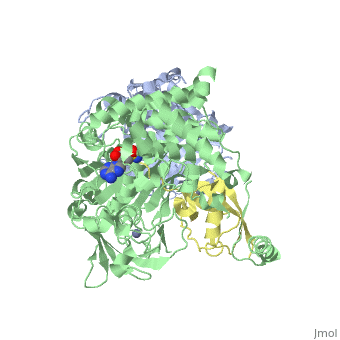
| - | <StructureSection load='3kyc' size=' | + | <StructureSection load='3kyc' size='350' scene='' caption='Human SUMO-1 (yellow) complex with SUMO-activating enzyme subunit 1 (grey), SUMO-activating enzyme subunit 2 (green), adenosine derivative and Zn+2 ion (grey) (PDB code [[3kyc]])'> |
| - | + | __TOC__ | |
| - | [[SUMO]] is a '''Small Ubiquitin-like MOdifier''' which covalently attaches to cellular proteins to modify their function. SUMO is similar in structure but not in sequence to [[Ubiquitin|ubiquitin]]. In several organisms SUMO is called SMT3. The SUMO-conjugating enzyme (E2) is called UBC9. The sentrin specific protease (SEPN) cleaves the C-terminal peptide from SUMO which then can bind to ubiquitin activating enzyme (E1). For details on SUMO-1 protein complex see <br /> | + | == Function == |
| + | [[SUMO]] is a '''Small Ubiquitin-like MOdifier''' which covalently attaches to cellular proteins to modify their function. SUMO is similar in structure but not in sequence to [[Ubiquitin|ubiquitin]]. In several organisms SUMO is called '''SMT3'''. The SUMO-conjugating enzyme (E2) is called UBC9. The sentrin specific protease (SEPN) cleaves the C-terminal peptide from SUMO which then can bind to ubiquitin activating enzyme (E1). For details on SUMO-1 protein complex see <br /> | ||
*[[Human SUMO E1 complex]] <br /> | *[[Human SUMO E1 complex]] <br /> | ||
*[[Human SUMO E1 complex with a SUMO1-AMP mimic]]<br /> | *[[Human SUMO E1 complex with a SUMO1-AMP mimic]]<br /> | ||
*[[Human SUMO E1~SUMO1-AMP tetrahedral intermediate mimic]]. | *[[Human SUMO E1~SUMO1-AMP tetrahedral intermediate mimic]]. | ||
| + | == Relevance == | ||
| + | Sumoylation may have a potential role in Alzheimer disease and decrease sumoylation of lamina A is a causative factor in familial dilated cardiomyopathy<ref>PMID:19282183</ref>. | ||
| + | |||
| + | == Structural highlights == | ||
[[Ubiquitin]] (Ub) and ubiquitin-like (Ubl) proteins attached to their target proteins and modulating the activities of those targets in various ways. Three types of evolutionarily conserved enzymes — E1 activating enzymes, E2 conjugating enzymes and E3 ligase enzymes — act sequentially through parallel yet distinct pathways to conjugate ubiquitin and Ubl proteins, such as SUMO and NEDD8, to their targets. The E1 enzyme uses the <scene name='3kyc/Cv/3'>adenosine triphosphate (ATP)</scene> and magnesium to adenylate the C-terminal Ub/Ubl glycine, releasing pyrophosphate and resulting in <scene name='3kyc/Cv/8'>adenosine monophosphate (AMP)</scene>. A non-hydrolysable <scene name='3kyc/Cv/4'>mimic of the acyl adenylate intermediate (AMSN)</scene> and <scene name='3kyc/Cv/5'>mimic of the tetrahedral intermediate (AVSN)</scene> were constructed. In both these compounds the atom of <font color='orange'><b>phosphorus</b></font> is replaced by sulfur (colored <font color='yellow'><b>yellow</b></font>). | [[Ubiquitin]] (Ub) and ubiquitin-like (Ubl) proteins attached to their target proteins and modulating the activities of those targets in various ways. Three types of evolutionarily conserved enzymes — E1 activating enzymes, E2 conjugating enzymes and E3 ligase enzymes — act sequentially through parallel yet distinct pathways to conjugate ubiquitin and Ubl proteins, such as SUMO and NEDD8, to their targets. The E1 enzyme uses the <scene name='3kyc/Cv/3'>adenosine triphosphate (ATP)</scene> and magnesium to adenylate the C-terminal Ub/Ubl glycine, releasing pyrophosphate and resulting in <scene name='3kyc/Cv/8'>adenosine monophosphate (AMP)</scene>. A non-hydrolysable <scene name='3kyc/Cv/4'>mimic of the acyl adenylate intermediate (AMSN)</scene> and <scene name='3kyc/Cv/5'>mimic of the tetrahedral intermediate (AVSN)</scene> were constructed. In both these compounds the atom of <font color='orange'><b>phosphorus</b></font> is replaced by sulfur (colored <font color='yellow'><b>yellow</b></font>). | ||
| - | The <scene name='3kyc/Al/2'>structural alignment</scene> of the crystal structures for human SUMO E1 in complex with SUMO adenylate (AMSN) and tetrahedral intermediate (AVSN) analogues revealed opened conformation (<font color='orange'><b>SUMO1 in orange</b></font>, <font color='blue'><b>SAE1 colored in blue</b></font>, and <font color='darkviolet'><b>other domains in darkviolet</b></font>) and closed conformation (<font color='yellow'><b>SUMO1 in yellow</b></font>, <font color='cyan'><b>SAE1 colored in cyan</b></font>, and <font color='magenta'><b>other domains in magenta</b></font>), respectively. In the <scene name='3kyc/Al/7'>open conformation</scene> ([[3kyc]]) the distance between Cys domain (including Cys173) and mimic of the acyl adenylate intermediate AMSN is very long, while in the <scene name='3kyc/Al/6'>closed conformation</scene> ([[3kyd]]), the catalytic Cys173 is posioned near AVSN and SUMO1, so the overall structure revealed dramatic rearrangement. This large conformational change forms the <scene name='3kyc/Al/8'>E1~SUMO1-AVSN tetrahedral intermediate analogue</scene>. | + | The <scene name='3kyc/Al/2'>structural alignment</scene> of the crystal structures for human SUMO E1 in complex with SUMO adenylate (AMSN) and tetrahedral intermediate (AVSN) analogues revealed opened conformation (<font color='orange'><b>SUMO1 in orange</b></font>, <font color='blue'><b>SAE1 colored in blue</b></font>, and <font color='darkviolet'><b>other domains in darkviolet</b></font>) and closed conformation (<font color='yellow'><b>SUMO1 in yellow</b></font>, <font color='cyan'><b>SAE1 colored in cyan</b></font>, and <font color='magenta'><b>other domains in magenta</b></font>), respectively. In the <scene name='3kyc/Al/7'>open conformation</scene> ([[3kyc]]) the distance between Cys domain (including Cys173) and mimic of the acyl adenylate intermediate AMSN is very long, while in the <scene name='3kyc/Al/6'>closed conformation</scene> ([[3kyd]]), the catalytic Cys173 is posioned near AVSN and SUMO1, so the overall structure revealed dramatic rearrangement. This large conformational change forms the <scene name='3kyc/Al/8'>E1~SUMO1-AVSN tetrahedral intermediate analogue</scene>.<ref>PMID:20164921</ref> |
| - | </ | + | |
| - | + | ||
| - | [[Image: | + | [[Image:Kyc_smaller.gif|275px|left|thumb]] |
| + | <br> | ||
| + | <br> | ||
For better understanding of the difference between these two conformations you can see this [[Morphs|morph]] (generated by using [http://polyview.cchmc.org/polyview3d.html POLYVIEW-3D: http://polyview.cchmc.org/polyview3d.html]; reload/refresh this page to restart this movie). Of note, in contrast to the previous figure, the same domains of these two structures ([[3kyc]] and [[3kyd]]) are colored in the same colors (<font color='yellow'><b>SUMO1 in yellow</b></font>, <font color='blue'><b>SAE1 colored in blue</b></font> and <font color='darkviolet'><b>other domains in darkviolet</b></font>). The catalytic Cys173 is shown in the spacefill representation and colored green, AMSN (or AVSN) are shown in the spacefill representation and colored in CPK colors. | For better understanding of the difference between these two conformations you can see this [[Morphs|morph]] (generated by using [http://polyview.cchmc.org/polyview3d.html POLYVIEW-3D: http://polyview.cchmc.org/polyview3d.html]; reload/refresh this page to restart this movie). Of note, in contrast to the previous figure, the same domains of these two structures ([[3kyc]] and [[3kyd]]) are colored in the same colors (<font color='yellow'><b>SUMO1 in yellow</b></font>, <font color='blue'><b>SAE1 colored in blue</b></font> and <font color='darkviolet'><b>other domains in darkviolet</b></font>). The catalytic Cys173 is shown in the spacefill representation and colored green, AMSN (or AVSN) are shown in the spacefill representation and colored in CPK colors. | ||
| - | {{Clear}} | ||
== 3D Structures of SUMO == | == 3D Structures of SUMO == | ||
| + | [[SUMO 3D Structures]] | ||
| - | + | </StructureSection> | |
| - | + | {{Clear}} | |
| - | + | ==Reference== | |
| + | <references/> | ||
| - | **[[2k8h]] – SUMO – NMR – ''Trypanosoma brucei''<br /> | ||
| - | **[[1u4a]] – hSUMO-3 (mutant) – NMR – human<br /> | ||
| - | **[[1a5r]] - hSUMO-1 - NMR<br /> | ||
| - | **[[1wm2]], [[1wm3]], [[2awt]], [[4bkg]], [[4npn]] – hSUMO-2<br /> | ||
| - | |||
| - | * SUMO+ubiquitin-like SUMO-conjugating enzyme | ||
| - | |||
| - | **[[2vrr]], [[2uyz]] – mSUMO-1+Ubc9<br /> | ||
| - | **[[2pe6]] - hSUMO-1+Ubc9<br /> | ||
| - | **[[3uip]], [[3uio]] - hSUMO-1+UBC9 + RAN GTPase-activating enzyme<br /> | ||
| - | **[[3uin]] - hSUMO-2+UBC9 + RAN GTPase-activating enzyme<br /> | ||
| - | |||
| - | * SUMO+sentrin specific protease | ||
| - | |||
| - | **[[2io0]] – pre-hSUMO-2+SEPN2<br /> | ||
| - | **[[2io1]] - pre-hSUMO-3+SEPN2<br /> | ||
| - | **[[2g4d]] - hSUMO-1+SEPN1<br /> | ||
| - | **[[2iy1]] - hSUMO-1+SEPN1 (mutant)<br /> | ||
| - | **[[2iyd]], [[2ckh]], [[3zo5]] - hSUMO-2+SEPN1<br /> | ||
| - | **[[1tgz]] - hSUMO-1+SEPN2<br /> | ||
| - | **[[2io3]] - pre-hSUMO-2+SEPN2 (mutant)+RAN GTPase-activating enzyme (mutant)<br /> | ||
| - | **[[2iy0]] - hSUMO-1+SEPN1 (mutant)+RAN GTPase-activating enzyme <br /> | ||
| - | |||
| - | * SUMO+ubiquitin-conjugating enzyme | ||
| - | |||
| - | **[[1z5s]] - hSUMO-1+E2+ RAN GTPase-activating enzyme<br /> | ||
| - | **[[2bf8]] - SUMO-1+E2 - bovine<br /> | ||
| - | |||
| - | * SMT3 | ||
| - | |||
| - | **[[2k1f]] – SMT3 – Fruit fly<br /> | ||
| - | **[[2eke]] – ySMT3+UBC9 – yeast<br /> | ||
| - | **[[1euv]] – ySMT3+ULP1 protease<br /> | ||
| - | **[[3v60]], [[3v61]] – ySMT3 + PCNA (mutant)<br /> | ||
| - | **[[3v62]] – ySMT3 + PCNA (mutant) + ATP-dependent DNA helicase SRS2<br /> | ||
| - | **[[3qht]] – ySMT3 + monobody<br /> | ||
| - | |||
| - | * SUMO+other proteins | ||
| - | |||
| - | **[[2asq]] – hSUMO-1+SUMO-binding motif in PIASX<br /> | ||
| - | **[[3kyc]], [[3kyd]] – hSUMO+SUMO-activating enzyme <br /> | ||
| - | **[[2rpq]] – hSUMO-3+activating transcription factor <br /> | ||
| - | **[[1wyw]] - hSUMO-1+thymine DNA glycosylate<br /> | ||
| - | **[[2d07]] - hSUMO-3+thymine DNA glycosylate<br /> | ||
| - | **[[2kqs]] - hSUMO-1+death domain-associated protein 6 fragment<br /> | ||
| - | **[[2las]] – hSUMO-1 + M-IR2 peptide<br /> | ||
| - | **[[4wjn]], [[4wjo]] – hSUMO-1 (mutant) + PML<br /> | ||
| - | **[[4wjp]], [[4wjq]] – hSUMO-1 (mutant) + DAXX<br /> | ||
| - | **[[3rzw]] – hSUMO-1 (mutant) + monobody<br /> | ||
| - | **[[2mp2]] – hSUMO-3 + E3 ubiquitin protein ligase<br /> | ||
| - | |||
| - | }} | ||
| - | ==Reference== | ||
| - | <ref group="xtra">PMID:20164921</ref><references group="xtra"/> | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
Reference
- ↑ Sarge KD, Park-Sarge OK. Sumoylation and human disease pathogenesis. Trends Biochem Sci. 2009 Apr;34(4):200-5. doi: 10.1016/j.tibs.2009.01.004. Epub, 2009 Mar 11. PMID:19282183 doi:http://dx.doi.org/10.1016/j.tibs.2009.01.004
- ↑ Olsen SK, Capili AD, Lu X, Tan DS, Lima CD. Active site remodelling accompanies thioester bond formation in the SUMO E1. Nature. 2010 Feb 18;463(7283):906-12. PMID:20164921 doi:10.1038/nature08765