Beta-glucosidase
From Proteopedia
(Difference between revisions)
| (19 intermediate revisions not shown.) | |||
| Line 3: | Line 3: | ||
{{Clear}} | {{Clear}} | ||
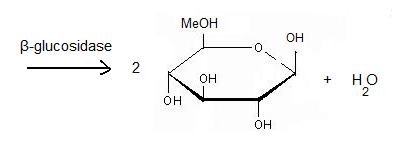
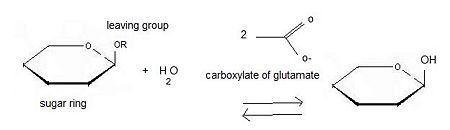
'''β-glucosidase''' is an enzyme which catalyses the hydrolysis of terminal non-reducing residues in β-glucosides (EC number : 3.2.1.21). In the case of 2VRJ, it comes from ''Thermotoga maritima'' which is a rod-shaped bacterium belonging to the order of Thermotogates. This bacterium was originally isolated from geothermal heated marine sediments. | '''β-glucosidase''' is an enzyme which catalyses the hydrolysis of terminal non-reducing residues in β-glucosides (EC number : 3.2.1.21). In the case of 2VRJ, it comes from ''Thermotoga maritima'' which is a rod-shaped bacterium belonging to the order of Thermotogates. This bacterium was originally isolated from geothermal heated marine sediments. | ||
| - | 2VRJ is here is in complex with an inhibitor called N-octyl-5-deoxy66-oxa-N-carbamoylcalystegine <ref>PMID: 18833549</ref>. | + | 2VRJ is here is in complex with an inhibitor called N-octyl-5-deoxy66-oxa-N-carbamoylcalystegine <ref>PMID: 18833549</ref>. |
| + | *'''Raucaffricine β-glucosidase''' (RGB) catalyzes the conversion of raucaffricine to glucose and vomilenine. | ||
| + | *'''6-phospho-glucosidase''' hydrolyzes maltose-6'-phosphate<ref>PMID: 15341727</ref>. | ||
| + | *'''Strictosidine-beta-glucosidase''' converts strictisidine to cathenamine<ref>PMID: 10652285</ref>. | ||
| + | *For '''Acid β-glucosidase''' see [[Acid-beta-glucosidase]]. | ||
| + | *For '''Glucan 1,3-glucosidase''' see [[Glucanase]]. | ||
| + | For Some more details see<br /> | ||
| + | * [[Molecular Playground/Beta-galactosidase]] | ||
| + | *[[Journal:Acta Cryst D:S205979832000501X|A Baculoviral System for the Production of Human β-Glucocerebrosidase Enables Atomic Resolution Analysis]] | ||
| + | *[[Beta-galactosidase (hebrew)]] | ||
===General action as biocatalyst=== | ===General action as biocatalyst=== | ||
| Line 54: | Line 63: | ||
The second step: '''hydrolysis''' is the most important. A cellulase is a complex of 3 enzymes which act together to hydrolyse cellulose: Endoglucanase breaks the chain in the middle of the molecular structure of cellulose. Exoglucanase binds an available end of the chain and isolates it. Then units of cellobiose are cut (two units of glucose which are together). To finish, '''β-glucosidase''' divides cellobiose into two glucoses. When they ferment, they become ethanol. The final product is obtained thanks to fermentation, distillation and deshydratation. | The second step: '''hydrolysis''' is the most important. A cellulase is a complex of 3 enzymes which act together to hydrolyse cellulose: Endoglucanase breaks the chain in the middle of the molecular structure of cellulose. Exoglucanase binds an available end of the chain and isolates it. Then units of cellobiose are cut (two units of glucose which are together). To finish, '''β-glucosidase''' divides cellobiose into two glucoses. When they ferment, they become ethanol. The final product is obtained thanks to fermentation, distillation and deshydratation. | ||
| + | == Treatment of Gaucher disease == | ||
| + | [http://en.wikipedia.org/wiki/Gaucher's_disease Gaucher disease], the most common [http://en.wikipedia.org/wiki/Lysosomal_storage_disease lysosomal storage disease], is caused by mutations in the gene that encoding the lysosomal enzyme, acid-β-glucosidase ([[acid-beta-glucosidase]], [http://en.wikipedia.org/wiki/Glucocerebrosidase glucocerebrosidase], GlcCerase, [http://www.expasy.org/cgi-bin/nicezyme.pl?3.2.1.45 E.C. 3.2.1.45]). The most common treatment for Gaucher disease is [http://en.wikipedia.org/wiki/Enzyme_replacement_therapy enzyme replacement therapy] (ERT), in which defective GlcCerase is supplemented with an active enzyme. | ||
| + | The correlation between the ~ 200 [http://en.wikipedia.org/wiki/Mutation mutations] in GlcCerase and disease severity is not completely understood, although [http://en.wikipedia.org/wiki/Zygosity#Homozygous homozygosity] for the common <scene name='1ogs/Mutations_n370_and_l444/2'>mutations N370S and L444P</scene> is associated with non-neuronopathic and neuronopathic disease, respectively. | ||
| + | |||
| + | ====Imiglucerase (Cerezyme®)==== | ||
| + | The [http://en.wikipedia.org/wiki/X-ray_crystallography X-ray structure] of GlcCerase ([http://en.wikipedia.org/wiki/Imiglucerase Cerezyme®]) was resolved at 2.0 A resolution ([[1ogs]]). The catalytic domain consists of a (beta/alpha)(8) TIM barrel, as expected for a member of the glucosidase hydrolase A family. The distance between the <scene name='1ogs/Catalytic_residues/2'>catalytic residues E235 and E340</scene> is consistent with a catalytic mechanism of retention. N370 is located on the longest alpha-helix (<scene name='1ogs/Helix_7/4'>helix 7</scene>), which has several other mutations of residues that point into the TIM barrel. Helix 7 is at the interface between the <scene name='1ogs/Tim1/2'>TIM barrel</scene> and a separate <scene name='1ogs/Ig_domain/2'>immunoglobulin-like domain</scene> on which L444 is located, suggesting an important regulatory or structural role for this non-catalytic domain. The structure provides the possibility of engineering improved GlcCerase for enzyme-replacement therapy, and for designing structure-based drugs aimed at restoring the activity of defective GlcCerase <ref name="Dvir">PMID:12792654</ref>. | ||
| + | |||
| + | {{Clear}} | ||
| + | ====GlcCerase with cyclohexitol==== | ||
| + | The crystal structure of the human <span style="color:yellow;background-color:black;font-weight:bold;">colored yellow</span> with covalently bound [http://en.wikipedia.org/wiki/Enzyme_inhibitor#Irreversible_inhibitors irreversible inhibitor] <scene name='1y7v/Bound_cyclohexitol/4'>cyclohexitol</scene> (<span style="color:cyan;background-color:black;font-weight:bold;">conduritol-B-epoxide; CBE; shown in cyan</span> with its <font color='red'><b>hydroxyl groups</b></font> are in <font color='red'><b>red</b></font>) was solved ([[1y7v]], <ref name="Premkumar">PMID:15817452</ref>). This structure reveals that binding of CBE to the active site does not induce a global conformational change in GlcCerase and confirms that Glu340 is the active-site catalytic nucleophile, because the <scene name='1y7v/Active_site1/3'>distance</scene> between the cyclohexitol C1 atom and Glu340 Oε2 is 1.43 Å. The comparison between the active sites of <scene name='1y7v/Active_site/13'>GlcCerase</scene> and another representative of the glycohydrolase family - plant <scene name='1y7v/Active_site_beta_glu_glyco/5'>β-D-glucan glucohydrolase</scene> ([[1iev]], <ref name="Hrmova">PMID:11709165</ref>), reveals that CBE bound with this plant enzyme adopted the "chair" conformation, while with human <scene name='1y7v/Active_site/14'>GlcCerase</scene>, it is observed in a "boat" conformation, with hydrogen bonds to Asn234 Oδ1 and Nδ2, Glu340 Oε1, Trp179 Nε1, and Asp127 Oδ1 and Oδ2 <ref name="Premkumar"/>. | ||
| + | Only one of two <scene name='1y7v/Loops/3'>alternative conformations</scene> of a pair of flexible loops (L1: Ser345–Glu349, and L2: Val394–Asp399) located at the entrance to the active site in native GlcCerase ([[1ogs]]) is observed in the GlcCerase-CBE structure ([[1y7v]]), a conformation in which the active site is accessible to CBE (<font color='blue'><b>colored blue</b></font>), while these loops in <font color='magenta'><b>the second (closed) conformation are colored magenta</b></font>. In <scene name='1y7v/L2/5'>loop 2</scene>, a major structural change is observed in the positions of <scene name='1y7v/L2/6'>Asn396 and Phe397</scene>, and in <scene name='1y7v/L1/6'>loop 1</scene> a more limited difference is observed in the conformations of <scene name='1y7v/L1/7'>Lys346 and Glu349</scene>. Analysis of the dynamics of these two alternative conformations suggests that the two loops act as a lid at the entrance to the active site. The movies [http://www.jbc.org/content/vol0/issue2005/images/data/M502799200/DC1/mov.mov 1] and [http://www.jbc.org/content/vol0/issue2005/images/data/M502799200/DC1/mov2.mov 2] illustrate the dynamics of the movement of these two loops <ref name="Premkumar"/><ref name="Zeev-Ben-Mordehai">PMID:12601798</ref>. | ||
| + | {{Clear}} | ||
| + | ====Native human acid β-glucosidase, expressed in cultured plant cells (prGCD, pGlcCerase)==== | ||
| + | Three-dimensional structure of recombinant plant-derived glucocerebrosidase (prGCD, [[2v3f]]) consists of <scene name='2v3f/Cv/7'>3 domains</scene>. <span style="color:pink;background-color:black;font-weight:bold;">Domain I (residues 1–27 and 384–414, colored pink)</span> comprises a 3-stranded anti-parallel β-sheet flanked by a perpendicular amino-terminal strand. <span style="color:lime;background-color:black;font-weight:bold;">Domain II (residues 30–75 and 431–497, colored lime)</span> consists of two β-sheets. <font color='red'><b>Domain III (residues 76–381 and 416–430, colored red)</b></font> is a (β/α) 8 TIM barrel. <scene name='2v3f/Cv/10'>The catalytic site</scene> with molecule BTB is shown <ref name="Shaaltiel">PMID:17524049</ref>. | ||
| + | <scene name='2v3f/Align/2'>Structural alignment</scene> of <font color='red'><b>prGCD</b></font> ([[2v3f]]) with both <span style="color:cyan;background-color:black;font-weight:bold;">Cerezyme®</span> ([[1ogs]]) and <span style="color:yellow;background-color:black;font-weight:bold;">Cerezyme® covalently modified by an irreversible inhibitor, conduritol-B-epoxide, colored yellow</span> ([[1y7v]]), revealed highly significant structural identity. The RMSD values for Cα atoms of these structures were of 0.64 and 0.60 Å, respectively. Moreover, there was strict conservation of the <scene name='2v3f/Align/3'>active site residues</scene> <ref name="Shaaltiel"/>. | ||
| + | |||
| + | {{Clear}} | ||
| + | ====pGlcCerase with ligands==== | ||
| + | <scene name='2v3d/Al/3'>Superimposition</scene> of the structure of <font color='red'><b>native human acid β-glucosidase, expressed in cultured plant cells (pGlcCerase,</b></font> [[2v3f]]) on those of <font color='darkmagenta'><b>N-butyl-deoxynojirimycin/pGlcCerase</b></font> ([[2v3d]]), <font color='lime'><b>N-nonyl-deoxynojirimycin/pGlcCerase</b></font> ([[2v3e]]), and <font color='cyan'><b>isofagomine/deglycosylated Cerezyme</b></font> (IFG/DG-Cerezyme, [[2nsx]]) reveals significant structural identity, neither of these ligands causes structural changes upon binding to the enzyme. The imino sugar of <font color='magenta'><b>N-butyl-deoxynojirimycin</b></font> <scene name='2v3d/Al/10'>(NB-DNJ)</scene> forms 7 hydrogen bonds and also makes several hydrophobic interactions with side chains of <font color='darkmagenta'><b>active site residues</b></font> ([[2v3d]]). The crystal structure of <font color='lime'><b>pGlcCerase in complex</b></font> with <font color='orange'><b>N-nonyl-deoxynojirimycin</b></font> <scene name='2v3d/Al/11'>(NN-DNJ)</scene> ([[2v3e]]) is very similar to that of <font color='magenta'><b>NB-DNJ</b></font>/<font color='darkmagenta'><b>pGlcCerase</b></font>. The exception is that longer chain of <font color='orange'><b>NN-DNJ</b></font> interacts with 2 additional residues Leu241 (<font color='lime'><b>labeled lime</b></font>) and Leu314 of symmetrically related monomer (not shown). Comparison of the structures of NB-DNJ/pGlcCerase ([[2v3d]]) and NN-DNJ/pGlcCerase ([[2v3e]]) with that of <scene name='2v3d/Nsx/2'>IFG/DG-Cerezyme</scene> ([[2nsx]]) shows that the pyranose-like ring forms a same number of hydrogen bonds with the enzyme in all three cases ([[2v3d]], [[2v3e]], and [[2nsx]]) <ref name="Shaaltiel"/><ref name="Brumshtein">PMID:17666401</ref><ref name="Lieberman">PMID:17187079</ref>. | ||
| + | {{Clear}} | ||
| + | ====Velaglucerase alfa==== | ||
| + | The <scene name='2wkl/Al/4'>structural alignment</scene> of the crystal structure of <font color='red'><b>velaglucerase alfa (colored red)</b></font> ([[2wkl]]) reveals that it is very similar to those of the recombinant GlcCerase produced in Chinese hamster ovary cells (<font color='blueviolet'><b>imiglucerase, Cerezyme®, colored blueviolet</b></font>, [[2j25]]) and in transgenic carrot cells (prGCD, [[2v3f]]). <scene name='2wkl/Al/13'>Superposition</scene> of the two individual molecules in the asymmetric unit of velaglucerase alfa and imiglucerase demonstrates striking similarity between positions of <font color='orange'><b>catalytic residues E235 and E340 (colored orange) in all 4 molecules</b></font>. The position of H311 is also very similar in all 4 molecules, whereas the conformations of 3 other active site residues W312, Y313, and, especially N396 are somewhat different. The active site residues (except <font color='orange'><b>E235 and E340</b></font>) of the two individual molecules in the asymmetric unit of velaglucerase alfa are colored: <font color='red'><b>subunit A (red)</b></font>, <font color='lime'><b>subunit B (lime)</b></font> and of imiglucerase: <font color='blueviolet'><b>subunit A (blueviolet)</b></font>, <font color='magenta'><b>subunit B (magenta)</b></font>. Imiglucerase and pr-GlcCerase contain a <scene name='2wkl/Al/14'>histidine</scene> at residue 495 <font color='blueviolet'><b>(blueviolet)</b></font>, whereas velaglucerase alfa contains <scene name='2wkl/Al/15'>arginine</scene> <font color='red'><b>(red)</b></font>. Mutations which cause Gaucher disease, <scene name='2wkl/Al/16'>R496 and D474</scene> are close to R495 near the N-terminus of GlcCerase. The <scene name='2wkl/Al/11'>velaglucerase alfa</scene> (<font color='blue'><b>its glycans are colored blue</b></font>) and <scene name='2wkl/Al/12'>imiglucerase</scene> (<font color='magenta'><b>its glycans are colored magenta</b></font>) have different carbohydrate composition <ref name="Shaaltiel"/><ref name="Wormald">PMID:17139081</ref><ref name="Salinas">PMID:19741058</ref>. This difference in glycosylation causes the increased cellular uptake of velaglucerase alfa over imiglucerase and could lead to improvement of treatment of Gaucher disease <ref name="Salinas"/>. | ||
==Additional Resources== | ==Additional Resources== | ||
For additional information, see: [[Carbohydrate Metabolism]] | For additional information, see: [[Carbohydrate Metabolism]] | ||
| - | </StructureSection> | ||
==3D structures of Beta-glucosidase== | ==3D structures of Beta-glucosidase== | ||
| + | [[Beta-glucosidase 3D structures]] | ||
| - | + | </StructureSection> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | **[[3zj7]], [[3zj8]] – seSGB + inhibitor<br /> | ||
| - | **[[2jf7]] - seSGB<br /> | ||
| - | **[[2jf6]] – seSGB + strictosidine<br /> | ||
| - | }} | ||
== References == | == References == | ||
<references/> | <references/> | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
References
- ↑ Aguilar M, Gloster TM, Garcia-Moreno MI, Ortiz Mellet C, Davies GJ, Llebaria A, Casas J, Egido-Gabas M, Garcia Fernandez JM. Molecular basis for beta-glucosidase inhibition by ring-modified calystegine analogues. Chembiochem. 2008 Nov 3;9(16):2612-8. PMID:18833549 doi:10.1002/cbic.200800451
- ↑ Rajan SS, Yang X, Collart F, Yip VL, Withers SG, Varrot A, Thompson J, Davies GJ, Anderson WF. Novel catalytic mechanism of glycoside hydrolysis based on the structure of an NAD+/Mn2+ -dependent phospho-alpha-glucosidase from Bacillus subtilis. Structure. 2004 Sep;12(9):1619-29. PMID:15341727 doi:10.1016/j.str.2004.06.020
- ↑ Geerlings A, Ibañez MM, Memelink J, van Der Heijden R, Verpoorte R. Molecular cloning and analysis of strictosidine beta-D-glucosidase, an enzyme in terpenoid indole alkaloid biosynthesis in Catharanthus roseus. J Biol Chem. 2000 Feb 4;275(5):3051-6. PMID:10652285 doi:10.1074/jbc.275.5.3051
- ↑ http://en.wikipedia.org/wiki/B-glucosidase
- ↑ Davies G, Henrissat B. Structures and mechanisms of glycosyl hydrolases. Structure. 1995 Sep 15;3(9):853-9. PMID:8535779
- ↑ http://www.ebi.ac.uk/interpro/IEntry?ac=IPR018120#PUB00002205
- ↑ http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/CSA/CSA_Site_Wrapper.pl?pdb=2vrj
- ↑ Davies G, Henrissat B. Structures and mechanisms of glycosyl hydrolases. Structure. 1995 Sep 15;3(9):853-9. PMID:8535779
- ↑ http://www.cazy.org/fam/ghf_INV_RET.html#3
- ↑ Dvir H, Harel M, McCarthy AA, Toker L, Silman I, Futerman AH, Sussman JL. X-ray structure of human acid-beta-glucosidase, the defective enzyme in Gaucher disease. EMBO Rep. 2003 Jul;4(7):704-9. PMID:12792654 doi:10.1038/sj.embor.embor873
- ↑ 11.0 11.1 11.2 Premkumar L, Sawkar AR, Boldin-Adamsky S, Toker L, Silman I, Kelly JW, Futerman AH, Sussman JL. X-ray structure of human acid-beta-glucosidase covalently bound to conduritol-B-epoxide. Implications for Gaucher disease. J Biol Chem. 2005 Jun 24;280(25):23815-9. Epub 2005 Apr 6. PMID:15817452 doi:M502799200
- ↑ Hrmova M, Varghese JN, De Gori R, Smith BJ, Driguez H, Fincher GB. Catalytic mechanisms and reaction intermediates along the hydrolytic pathway of a plant beta-D-glucan glucohydrolase. Structure. 2001 Nov;9(11):1005-16. PMID:11709165
- ↑ Zeev-Ben-Mordehai T, Silman I, Sussman JL. Acetylcholinesterase in motion: visualizing conformational changes in crystal structures by a morphing procedure. Biopolymers. 2003 Mar;68(3):395-406. PMID:12601798 doi:10.1002/bip.10287
- ↑ 14.0 14.1 14.2 14.3 Shaaltiel Y, Bartfeld D, Hashmueli S, Baum G, Brill-Almon E, Galili G, Dym O, Boldin-Adamsky SA, Silman I, Sussman JL, Futerman AH, Aviezer D. Production of glucocerebrosidase with terminal mannose glycans for enzyme replacement therapy of Gaucher's disease using a plant cell system. Plant Biotechnol J. 2007 Sep;5(5):579-90. Epub 2007 May 24. PMID:17524049 doi:10.1111/j.1467-7652.2007.00263.x
- ↑ Brumshtein B, Greenblatt HM, Butters TD, Shaaltiel Y, Aviezer D, Silman I, Futerman AH, Sussman JL. Crystal structures of complexes of N-butyl- and N-nonyl-deoxynojirimycin bound to acid beta-glucosidase: insights into the mechanism of chemical chaperone action in Gaucher disease. J Biol Chem. 2007 Sep 28;282(39):29052-8. Epub 2007 Jul 31. PMID:17666401 doi:10.1074/jbc.M705005200
- ↑ Lieberman RL, Wustman BA, Huertas P, Powe AC Jr, Pine CW, Khanna R, Schlossmacher MG, Ringe D, Petsko GA. Structure of acid beta-glucosidase with pharmacological chaperone provides insight into Gaucher disease. Nat Chem Biol. 2007 Feb;3(2):101-7. Epub 2006 Dec 24. PMID:17187079 doi:http://dx.doi.org/10.1038/nchembio850
- ↑ Brumshtein B, Wormald MR, Silman I, Futerman AH, Sussman JL. Structural comparison of differently glycosylated forms of acid-beta-glucosidase, the defective enzyme in Gaucher disease. Acta Crystallogr D Biol Crystallogr. 2006 Dec;62(Pt 12):1458-65. Epub 2006, Nov 23. PMID:17139081 doi:S0907444906038303
- ↑ 18.0 18.1 Brumshtein B, Salinas P, Peterson B, Chan V, Silman I, Sussman JL, Savickas PJ, Robinson GS, Futerman AH. Characterization of gene-activated human acid-beta-glucosidase: crystal structure, glycan composition, and internalization into macrophages. Glycobiology. 2010 Jan;20(1):24-32. Epub 2009 Sep 9. PMID:19741058 doi:10.1093/glycob/cwp138
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Muriel Breteau, Alexander Berchansky, Joel L. Sussman, David Canner