We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Flagellar proteins
From Proteopedia
(Difference between revisions)
(New page: <StructureSection load='5h5v' size='340' side='right' caption='E. coli FliD D1-D2-D3 domain (PDB entry 1p6o)' scene=''> == Function == The '''Flagelar hook-associated proteins FlgK...) |
m (Flagellar hook-associated protein moved to Flagellar proteins: requested by Editor) |
||
| (6 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
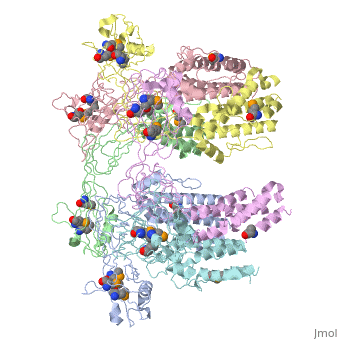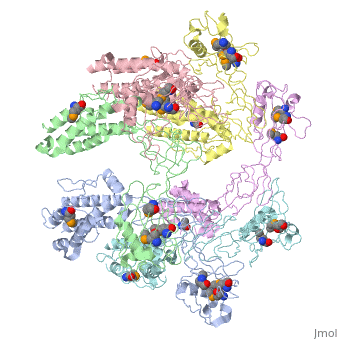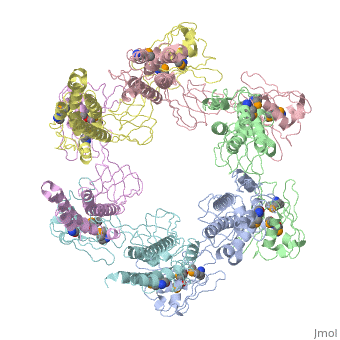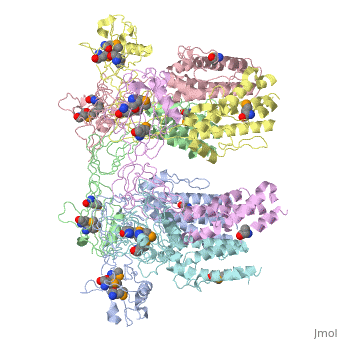
| - | + | <StructureSection load='5h5v' size='340' side='right' caption='E. coli Se-Met FliD D1-D2-D3 domain (PDB entry [[5h5v]])' scene=''> | |
| - | <StructureSection load='5h5v' size='340' side='right' caption='E. coli FliD D1-D2-D3 domain (PDB entry [[ | + | |
== Function == | == Function == | ||
| - | The ''' | + | The '''Flagellar proteins FlgK''' (or HAP1), '''FlgL''' (or HAP3) and '''FliD''' (or HAP2) are part of the bacterial flagellum <ref>PMID:2193164</ref>. These 3 proteins seem to be located at the claw-shaped end of the flagellum and play an essential role in filament formation. '''Flagellar protein FlgA''' is a chaperone protein required for P-ring assembly in bacterial flagella<ref>PMID:27273476</ref>. For details see [[Samatey/2]]. |
</StructureSection> | </StructureSection> | ||
| - | == 3D Structures of | + | == See Also == |
| + | * [[Flagella, bacterial]] | ||
| + | * [[Flagellar filament of bacteria]] | ||
| + | * [[The Bacterial Flagellar Hook]] | ||
| + | * [[Samatey/4|Complete Flagellar Hook Structure]] determined by cryo-EM in 2016. | ||
| + | * [[Samatey/3|Structural insights into bacterial flagellar hook similarities and specifities]] (2016). | ||
| + | * Other recent reports on structures of flagellar components at [[Samatey]]. | ||
| + | |||
| + | == 3D Structures of flagellar proteins == | ||
Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| Line 22: | Line 29: | ||
[[5xlk]], [[5xlj]] – FliD D2-D3 domain – ''Serratia marcascens''<br /> | [[5xlk]], [[5xlj]] – FliD D2-D3 domain – ''Serratia marcascens''<br /> | ||
[[5fhy]] – FliD D2-D3 domain – ''Pseudomonas aeruginosa''<br /> | [[5fhy]] – FliD D2-D3 domain – ''Pseudomonas aeruginosa''<br /> | ||
| + | [[3vki]], [[3vjp]], [[3tee]] – StFlgA <br /> | ||
| + | [[3frn]] – FlgA – ''Thermotoga maritima''<br /> | ||
== References == | == References == | ||
Current revision
| |||||||||||
See Also
- Flagella, bacterial
- Flagellar filament of bacteria
- The Bacterial Flagellar Hook
- Complete Flagellar Hook Structure determined by cryo-EM in 2016.
- Structural insights into bacterial flagellar hook similarities and specifities (2016).
- Other recent reports on structures of flagellar components at Samatey.
3D Structures of flagellar proteins
Updated on 01-January-2018
4ut1 – FlgK – Burkholderia pseudomallei
2d4x – StFlgL – Salmonella typhimurium
2d4y – StFlgK
5gna – StFlgK residues 401-467 + FliT
5h5t – StFliD D2-D3 domain
5h5v – EcFliD D1-D2-D3 domain – Escherichia coli
5h5w – EcFliD D2-D3 domain
5xlk, 5xlj – FliD D2-D3 domain – Serratia marcascens
5fhy – FliD D2-D3 domain – Pseudomonas aeruginosa
3vki, 3vjp, 3tee – StFlgA
3frn – FlgA – Thermotoga maritima
References
- ↑ Homma M, DeRosier DJ, Macnab RM. Flagellar hook and hook-associated proteins of Salmonella typhimurium and their relationship to other axial components of the flagellum. J Mol Biol. 1990 Jun 20;213(4):819-32. PMID:2193164
- ↑ Matsunami H, Yoon YH, Meshcheryakov VA, Namba K, Samatey FA. Structural flexibility of the periplasmic protein, FlgA, regulates flagellar P-ring assembly in Salmonella enterica. Sci Rep. 2016 Jun 7;6:27399. doi: 10.1038/srep27399. PMID:27273476 doi:http://dx.doi.org/10.1038/srep27399