From Proteopedia
(Difference between revisions)
proteopedia linkproteopedia link
|
|
| Line 11: |
Line 11: |
| | | | |
| | ==Structure== | | ==Structure== |
| - | ===General Structure===
| + | <table style="width: 100%;"> |
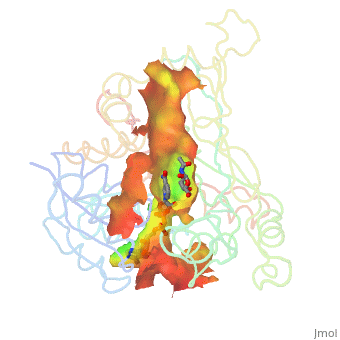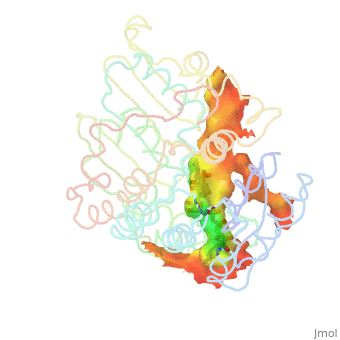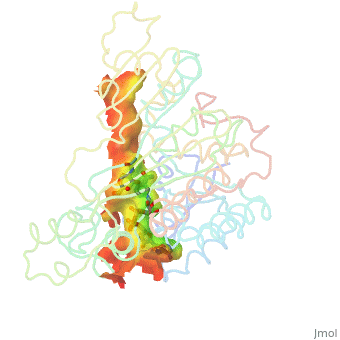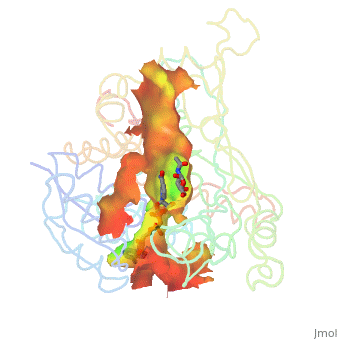
| - | There are two distinct classes of HMGRs, class I, which is only found in eukaryotes and are membrane bound and class II, which is found in prokaryotes and are soluble. <ref>PMID:11349148</ref> HMGR contains 8 transmembrane domains, which have yet to be successfully crystallized, that anchor the protein to the membrane of the endoplasmic reticulum. <ref name="Roitelman">PMID:1374417</ref> The catalytic portion of human HMGR forms a tetramer, with the individual monomers winding around each other. <ref name="Roitelman"/> Within the tetramer, the monomers are arranged into <scene name='HMG-CoA_Reductase/1dq8_2_dimers/1'>two dimers</scene>, each of which contains <scene name='HMG-CoA_Reductase/1dq8_2_active_sites/1'>two active sites </scene>which are formed by residues form both monomers. Each monomer contains <scene name='HMG-CoA_Reductase/1dq8_star3_domains/1'>three domains </scene>, the <scene name='HMG-CoA_Reductase/1dq8_n_domain/2'>N-domain</scene>, the <scene name='HMG-CoA_Reductase/1dq8_l_domain/1'>L-Domain</scene>, and the <scene name='HMG-CoA_Reductase/1dq8_s_domain/1'>S-Domain</scene>. The L-domain is unique to HMGRs while the S-domain, which forms the binding site for NADP, resembles that of [[ferredoxin]]. The S and L domains are connected by a <scene name='HMG-CoA_Reductase/1dq8_cis_loop/1'>“cis-loop”</scene> which is essential for the HMG-binding site. <ref name="Roitelman"/> Salt bridges between residues R641 and E782 as well as <scene name='HMG-CoA_Reductase/1dq8_cis_loop/2'>hydrogen bonds</scene> between E700 and E700 on neighboring monomers compliment the largely hydrophobic dimer-dimer interface. <ref name="Roitelman"/>
| + | <tr> |
| - | <br /> | + | <td> |
| - | | + | <applet load='1aoi' size='600' frame='true' align='right' caption='' /> |
| - | | + | </td> |
| - | ===Substrate Binding & Catalytic Mechanism===
| + | <td> |
| - | <applet load="1dqa2.pdb" size="400" color="white" frame="true" spin="on" Scene ="HMG-CoA_Reductase/1dqa_starting_2/1" caption="Crystal Structure of Human HMGR, [[1dqa]]" align="right"/> | + | <div style="background-color: #ffe8e8;overflow: auto;height: 600px;width: 100%;"> |
| - | [[Image: Reactin_scheme.PNG|300px|left|thumb| Chemical Reaction Catalyzed by HMGR]]
| + | {{:User:Eran Hodis/Sandbox8}} |
| - | The HMG-CoA and NADPH molecules make numerous contacts with the L and S domains in forming the four active sites. The CoA is located in a <scene name='HMG-CoA_Reductase/1dqa_nadp_and_coa/1'>positively charged pocket near the enzyme surface</scene>, with the pantothenic acid moiety extending into the interior of the protein. <scene name='HMG-CoA_Reductase/1dqa_tyr_491/2'>Tyrosine 479 forms a hydrophobic lid</scene> over the CoA adenine base, shielding the extended binding pocket from solution. The NADPH binding site is formed primarily by the S-domain with <scene name='HMG-CoA_Reductase/1dqa_loop/2'>a loop region</scene> playing a critical role in binding. <ref name="Roitelman"/>
| + | </div> |
| - | | + | </td> |
| - | The HMG binding pocket is the site of catalysis in HMGR. <scene name='HMG-CoA_Reductase/1dqa_cis_loop2/2'> The“cis-loop” that bends over the top of HMG </scene> is a critical structural element of this binding site. Residues <scene name='HMG-CoA_Reductase/1dqa_e_and_d/2'>E559 and D767</scene> and are positioned in the active site as is <scene name='HMG-CoA_Reductase/1dqa_k691/2'>K691 which is only 2.7 angstroms from the HMG O2 carbonyl oxygen</scene>. It is this K691 that presumably stabilizes the negatively charged oxygen on the first mevaldyl-CoA intermediate. <ref name="Roitelman"/> The mevaldyl CoA intermediate is subsequently converted to Mavaldehyde with added stabilization from <scene name='HMG-CoA_Reductase/1dqa_h866/2'>H866, which is within hydrogen bonding distance of the thiol group</scene>. It is then believed that the close proximity of <scene name='HMG-CoA_Reductase/1dqa_e_and_d/2'>E559 and D767</scene> increases the pKA of E559, allowing it to be a proton donor for the reduction of mevaldehyde into mevalonate. <ref name="Roitelman"/>
| + | </tr> |
| - | <br /> | + | </table> |
| | | | |
| | ==Regulation of HMG-CoA Reductase== | | ==Regulation of HMG-CoA Reductase== |
Revision as of 14:40, 29 October 2010
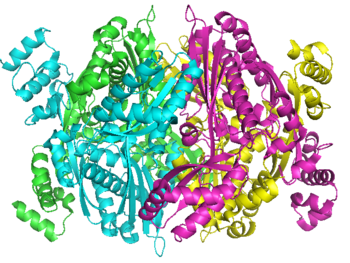
Human HMG-CoA Reductase Catalytic Domain,
1dqa
|
|
| Crystal Structure of Human HMG-CoA Reductase Catalytic Domain, 1dq8
|
| Ligands:
| , ,
|
| Activity:
| Hydroxymethylglutaryl-CoA reductase (NADPH), with EC number 1.1.1.34
|
| Structural annotation:
|
| Resources:
| CATH : 1Dq8A03, 1Dq8A02, 1Dq8A01, 1Dq8B02, 1Dq8B01, 1Dq8B03, 1Dq8C02, 1Dq8C01, 1Dq8C03, 1Dq8D03, 1Dq8D02, 1Dq8D01
InterPro : Ipr009023, Ipr000731, Ipr002202, Ipr004816, Ipr004554, Ipr009029
Pfam : PF00368
SCOP : d1dq8a4, d1dq8a1, d1dq8b4, d1dq8b1, d1dq8c1, d1dq8c4, d1dq8d1, d1dq8d4
UniProt : P04035
|
|
|
|
|
|
| Resources:
| FirstGlance, OCA, RCSB, PDBsum
|
| Coordinates:
| save as pdb, mmCIF, xml
|
HMG-CoA Reductase (or 3-hydroxy-3-methyl-glutaryl-CoA reductasese or HMGR) is the rate-controlling enzyme of the metabolic pathway responsible for cholesterol and other isoprenoid biosynthesis, the mevalonate pathway. HMGR is a polytopic, transmembrane protein, containing 8 domains, that is anchored in the membrane of the endoplasmic reticulum.[1] It is the major target of the statins, a cholesterol lowering drug class and the best selling pharmaceutical drugs in the world.
Biological Role
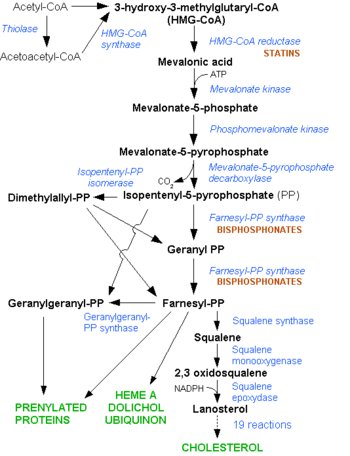
Mevalonate Pathway. Note the early stage at which the Statins interfere in the pathway
HMGR is among the most highly regulated enzymes in the human body. It catalyzes the formation of mevalonic acid, the committed step in the biosynthesis of sterols, notably cholesterol and isoprenoids. This reaction can be seen below where HMG-CoA is reduced by NADPH. Despite the poor reputation cholesterol has in the media, it is a critical component of cellular membranes as it is required to establish proper membrane permeability and fluidity. The mevalonate pathway is also responsible for synthesis of the oxygen transporting heme found in red blood cells. [2]
Structure
|
|
|
Welcome to Proteopedia
The free, collaborative 3D-encyclopedia of proteins & other molecules
|
|
|
|
|
Featured Article
|
Green links change the 3D image!
Click and drag on the molecule!
|
|
|
|
|
|
The X-ray structure of HIV-1 protease reveals that it is composed of , each consisting of 99 amino acid residues. The subunits come together in such as way as to . This tunnel is of critical importance because the active site of the protease is located in its interior. The active site consists of , making it a member of the aspartyl protease family. The two Asp's are either interact with the incoming water OR protonate the carbonyl to make the carbon more electrophilic for the incoming . (more...)
|
Browse Proteopedia
|
|
Find Your Protein or Biomolecule
|
|
- All PDB entries (over 100,000) have pages
- Go takes you directly to the page if it exists,
- Search gives you search results
|
|
|
What Can Proteopedia Do For You?
|
|
|
|
|
Proteopedia News
|
How to display cavities, pockets, and tunnels. |
- March 2024: Professors are using Proteopedia for class projects in Brazil, Czech Republic, France, and various states in the USA. See the updated Adoptions in College and University Classes.
- What happens if a SARS-CoV-2 coronavirus enters your lung? See a clear explanation at Lifecycle of SARS-CoV-2
- July 2022: Exercise-induced N-lactoyl-phenylalanine, appetite and obesity
- 5,000 users!! On December 4, 2021, the number of Proteopedia users went over 5,000. The 5,000th user is from Juniata College, Huntingdon, Pennsylvania, US
- AlphaFold protein structure predictions - a step change for biology, an electronic talk by EBI staff for students and early career researchers via FEBS Junior Sections, Oct. 26, 2021, 19:00 CEST. Announcement. Registration.
- A practical guide to teaching with Proteopedia [1]
- January 2021: How to display cavities, pockets, and tunnels? Jmol/Cavities_pockets_and_tunnels
- December 2020: What is changing on SARS-CoV-2 virus and what this means for humanity? SARS-CoV-2 spike protein mutations.
- August 2020: An animation of coronavirus spike protein showing the SARS-CoV-2 spike protein fusion transformation.
- July 2020: An animation of coronavirus spike protein showing SARS-CoV-2 protein S priming by furin.
- March 2020: New Proteopedia page focused on the Coronavirus 2019 (COVID-19).
- January 2019: New morphing feature for Proteopedia, powered by PyMOL from from Schrodinger.
- January, 2018: 10th Anniversary Celebration Conference, University of Massachusetts, Amherst, USA. Group Photo of Participants.
- September, 2017: A workshop based on Proteopedia was held at the New Horizons in Biochemistry & Molecular Biology Education Conference, jointly organised by FEBS and IUBMB and hosted at the Weizmann Institute of Science.
- June, 2016: Animate any Proteopedia scene in Powerpoint.
- June, 2016: Proteopedia uses the Biological Assemblies from PDBe as the default scenes for all PDB entry pages. Thus, based on the curation by EBI (host for PDBe) the most biologically significant structure is shown.
- June, 2015: Award Ceremony for the the Proteopedia Award at the ICSG2015 - Deep Sequencing Meets Structural Biology, awarded on 10-Jun-2015 at the Weizmann Institute of Science
- December, 2014, Talking about Proteopedia on 12/04/14-12/06/14, a live online talk organised by DivCHED CCCE: Committee on Computers in Chemical Education
- October, 2014 Course in Spanish/English on Proteopedia and its uses to study, display and teach macromolecules.
- How to create fast loading pages in Proteopedia.
- How to be as safe as possible with Java
- Proteopedia on iPads!
- What version of Jmol is running?
- Proteopedia status
- Awards
- ↑ Castro C, Johnson RJ, Kieffer B, Means JA, Taylor A, Telford J, Thompson LK, Sussman JL, Prilusky J, Theis K. A practical guide to teaching with Proteopedia. Biochem Mol Biol Educ. 2021 Jun 3. doi: 10.1002/bmb.21548. PMID:34080750 doi:http://dx.doi.org/10.1002/bmb.21548
|
The Scoreboard
|
Score based on pages-edited and number-of-edits,
and excludes members of the Proteopedia Team
All scores...
|
Want To Contribute?
|
Pages are easy to create and edit,
and green links are fun & easy to make!
Step 1:
Step 2:
Step 3:
|
Read The Article
|
|
|
|
|
| contact@proteopedia.org
|
|
|
Regulation of HMG-CoA Reductase
As stated before, HMGR is among the most highly regulated enzymes in the human body. [2] Regulation of HMG-CoA reductase occurs at 4 different levels of feedback regulation. First by regulation of transcription of the reductase gene, which is activated by sterol regulatory element binding protein, a protein that binds to the promoter of the HMGR gene when cholesterol levels fall. The second level of regulation is at the translation of the HMGR mRNA, which is inhibited by Farnesol, a derivative of the mevalonate pathway. [2]
The third level of HMGR regulation involves the degradation of intact reductase. As the level of sterols increases, HMGR reductase becomes more susceptible to ER-associated degradation. Helices 2-6 of the HMGR transmembrane domain, called the sterol-sensing domain, sense the increased levels of sterols. Degradation of HMGR is initiated when a membrane-bound cysteine protease cleaves within the transmembrane region of HMGR. Additionally, as sterol levels increase, the helices can slightly rearrange, exposing Lysine 248 which can be ubiquitinated and subsequently trigger proteolytic degradation. [1] [3]
A final level of HMGR regulation is achieved by inhibition via . HMGR is phosphorylated and inactivated by an AMP-activated protein kinase, when the energy charge of the cell is low and AMP concentrations are high. Since Serine 872 is in the vicinity of the pohsphate of NADP, phosphorylation likely results in a decreased affinity for NADPH, halting the formation of Mevaldyl-CoA from HMG-CoA.
[4]
Medical Implications
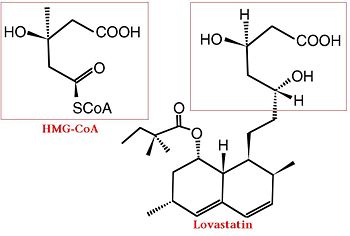
Comparison of Chemical Structure of HMG-CoA and Lovastatin
Elevated cholesterol levels have been identified as a major risk factor for coronary artery disease, the narrowing of arteries of the heart, which affected over 13 million people in the United States alone. It is a major cause of disability and death, killing over 500 thousand people in the USA in 2001. [5]
The statins are HMG-CoA reductase inhibitors. Discovered by Akira Endo in 1971 [6], stains are similar in structure to HMG-CoA and act by competitively inhibiting HMGR. Since HMGR is the first committed enzyme in the cascade that eventually produces cholesterol, use of statins can dramatically reduce blood cholesterol levels. As a drug class, statins generated over $20 billion dollars in sales in 2009 with Pfizer’s lipitor the best selling drug in the world. [7][8]
A number of crystal structures of HMGR with bound statins have been solved which elucidate how the statin molecule is bound by HMGR. Statins in general occupy the active site of HMGR, preventing HMG-CoA from being bound by the enzyme. The structures of HMGR with bound , , , , , and highlight the important residues involved in inhibitor binding. The cis loop forms a number of polar interactions with the inhibitor, particularly residues Ser 684, Asp 690, Lys 691, Lys 692, and hydrogen bond interactions between Glu 559 and Asp 767 with the O5-hydroxyl of the statins. Van der Waals interactions between Leu 562, Val 683, Leu 853, Ala 856, and Leu 857 of HMGR and hydrophobic ring structures of the statins. [9]These interactions serve to allow the statins to be bound by HMGR with a Ki of between .1-2.3nM while the Michaelis constant for HMG-CoA is 4uM. [10]
Additional 3D Structures of HMG-CoA Reductase
3cct, 3ccw, 3ccz, 3cd0, 3cd5, 3cd7, 3cda, 3cdb – HMG-CoA Reductase Catalytic Domain
2qil – HMG-CoA Reductase Catalytic Domain
2q6b, 2q6c – Catalytic Domain of HMG-CoA Reductase
1t02 – Crystal Structure of Statin Bound HMG-CoA Reductase
1r31 – Crystal Structure of HMGR from Pseudomonas mevalonii
1hw8, 1hw9, 1hwi, 1hwj, 1hwk, 1hwl – HMG CoA Reductase with Various Statins Bound
1qax, 1qay – Ternary Complex of Pseudomonas Mevalonii HMGR with mevalonate
Additional Resources
References
- ↑ Cite error: Invalid
<ref> tag;
no text was provided for refs named Roitelman
- ↑ 2.0 2.1 2.2 Meigs TE, Roseman DS, Simoni RD. Regulation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase degradation by the nonsterol mevalonate metabolite farnesol in vivo. J Biol Chem. 1996 Apr 5;271(14):7916-22. PMID:8626470
- ↑ Song BL, Sever N, DeBose-Boyd RA. Gp78, a membrane-anchored ubiquitin ligase, associates with Insig-1 and couples sterol-regulated ubiquitination to degradation of HMG CoA reductase. Mol Cell. 2005 Sep 16;19(6):829-40. PMID:16168377 doi:10.1016/j.molcel.2005.08.009
- ↑ Goldstein JL, Brown MS. Regulation of the mevalonate pathway. Nature. 1990 Feb 1;343(6257):425-30. PMID:1967820 doi:http://dx.doi.org/10.1038/343425a0
- ↑ www.nhlbi.nih.gov/health/.../Diseases/.../CAD_WhatIs.html
- ↑ Endo A, Kuroda M, Tanzawa K. Competitive inhibition of 3-hydroxy-3-methylglutaryl coenzyme A reductase by ML-236A and ML-236B fungal metabolites, having hypocholesterolemic activity. FEBS Lett. 1976 Dec 31;72(2):323-6. PMID:16386050
- ↑ http://www.drugs.com/top200.html
- ↑ http://www.medicalnewstoday.com/articles/25046.php
- ↑ Istvan ES, Deisenhofer J. Structural mechanism for statin inhibition of HMG-CoA reductase. Science. 2001 May 11;292(5519):1160-4. PMID:11349148 doi:10.1126/science.1059344
- ↑ Corsini A, Maggi FM, Catapano AL. Pharmacology of competitive inhibitors of HMG-CoA reductase. Pharmacol Res. 1995 Jan;31(1):9-27. PMID:7784310