Methyl CpG Binding Protein 2
From Proteopedia
(Difference between revisions)
| Line 2: | Line 2: | ||
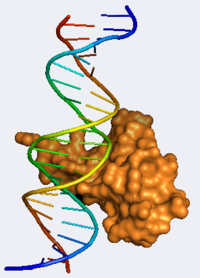
[[Image:Mecp2 picture.png|200px|left]] [[Methyl CpG Binding Protein 2]] ('''MeCP2''') is a transcriptional repressor that plays a critical role in mediating gene silencing. It binds the methylated BDNF [[DNA]] sequence with exquisite specificity while recruiting repressor complexes which include chromatin-remodeling enzymes.<ref name="Bird"/> MeCP2 has been found in most tissues but is highly concentrated in cortical neurons where it is believed to help modulate neuronal maturity and plasticity.<ref>PMID:14593168</ref> It is believed to play a crucial role in synaptogenesis and in the maintenance of neuronal function.<ref name="Bird">PMID:18313390</ref> Of particular note, mutations within the MeCP2 gene are responsible for over 95% of cases of Rett Syndrome. Rett Syndrome (RTT) is an [[Autism|autism]] spectrum neurodevelopmental disorder caused by the expression of mutant copies of the X-linked MeCP2 gene in neurons. Patients with RTT show abnormal neuronal morphology and have a large spectrum of autistic characteristics such as impaired motor function and regression of developmental skills, typically appearing 6-18 months after birth.<ref>PMID:21074045</ref> Intriguingly, Guy et al. demonstrated that the restoration of neuronal function by late expression of MeCP2 in mice can reverse many of the phenotypic traits associated with RTT.<ref>PMID:17289941</ref> | [[Image:Mecp2 picture.png|200px|left]] [[Methyl CpG Binding Protein 2]] ('''MeCP2''') is a transcriptional repressor that plays a critical role in mediating gene silencing. It binds the methylated BDNF [[DNA]] sequence with exquisite specificity while recruiting repressor complexes which include chromatin-remodeling enzymes.<ref name="Bird"/> MeCP2 has been found in most tissues but is highly concentrated in cortical neurons where it is believed to help modulate neuronal maturity and plasticity.<ref>PMID:14593168</ref> It is believed to play a crucial role in synaptogenesis and in the maintenance of neuronal function.<ref name="Bird">PMID:18313390</ref> Of particular note, mutations within the MeCP2 gene are responsible for over 95% of cases of Rett Syndrome. Rett Syndrome (RTT) is an [[Autism|autism]] spectrum neurodevelopmental disorder caused by the expression of mutant copies of the X-linked MeCP2 gene in neurons. Patients with RTT show abnormal neuronal morphology and have a large spectrum of autistic characteristics such as impaired motor function and regression of developmental skills, typically appearing 6-18 months after birth.<ref>PMID:21074045</ref> Intriguingly, Guy et al. demonstrated that the restoration of neuronal function by late expression of MeCP2 in mice can reverse many of the phenotypic traits associated with RTT.<ref>PMID:17289941</ref> | ||
| + | ====Structure of MeCP2==== | ||
The crystal structure of the DNA binding domain of MeCP2 was solved by Ho et al. in 2008 providing researchers with critical new information about the cause of RTT at the molecular level. <scene name='Methyl_CpG_Binding_Protein_2/Protein/3'>The MeCP2 DNA Binding Domain (MBD)</scene> binds the <scene name='Methyl_CpG_Binding_Protein_2/Dna/3'>methylated BDNF DNA sequence</scene> using a predominantly <scene name='Methyl_CpG_Binding_Protein_2/Hydrophil/1'>hydrophilic pocket</scene>. MeCP2 recognition of the mCpG sequence involves <scene name='Methyl_CpG_Binding_Protein_2/Water/1'>five water molecules</scene> each making CH-O hydrogen bonds. <scene name='Methyl_CpG_Binding_Protein_2/Water_1/1'>W22 forms hydrogen bonds</scene> with Asp 121, W24, W21, and N4 of m5C33. <scene name='Methyl_CpG_Binding_Protein_2/Water_2/3'>W24 forms hydrogen bonds</scene> with Tyr 123, Arg 133, water 22, and both N4 of m5C8 and a CH-O interaction with the methyl group of m5C8. The only residues that directly interact with DNA bases <scene name='Methyl_CpG_Binding_Protein_2/Asp/1'>are Asp 121</scene>, forming a CH-O hydrogen bond with methyl of m5C8, <scene name='Methyl_CpG_Binding_Protein_2/Fingers/1'>Arg 111, and Arg 133</scene>, each of which form symmetrical hydrogen bonds with each guanine in the mCpG pair. Both of these “Arginine Fingers” <scene name='Methyl_CpG_Binding_Protein_2/Coplanar/2'>lie in a plane with the guanine bases</scene> and are locked into position by <scene name='Methyl_CpG_Binding_Protein_2/Salt_bridges/1'>salt bridges with Asp 121 and Glu 137</scene>, placing the gaunidinum groups directly <scene name='Methyl_CpG_Binding_Protein_2/Methylated/1'>above/below the methyl groups </scene> of the methylated cytidine bases.<ref name="Bird"/> The C-terminal region of <scene name='Methyl_CpG_Binding_Protein_2/Mbd/1'>the MBD</scene> includes an unusual <scene name='Methyl_CpG_Binding_Protein_2/Tandem/1'>tandem Asx-ST motif</scene>, that consists of an Asx turn (Residues 156-158) followed by an ST motif (158-161). The <scene name='Methyl_CpG_Binding_Protein_2/Asx/1'>Asx turn is formed</scene> by a hydrogen bond that connects the main chain nitrogen of Thr 158 and the side chain of Asp 156, while the <scene name='Methyl_CpG_Binding_Protein_2/St/2'>ST motif is held together </scene> by hydrogen bonds to Thr 158. T158M, which is the most common missense mutation causing Rett Syndrome abolishes DNA binding because it disrupts this Asx-ST motif. Another well-known mutation, causing RTT, R106W, disrupts the motif stabilizing hydrogen bonds formed between <scene name='Methyl_CpG_Binding_Protein_2/106/2'>Arg 106 and Thr 158 and Val 159</scene>. This Asx-ST motif stabilizes MeCP2’s interaction with DNA by specifically binding to the <scene name='Methyl_CpG_Binding_Protein_2/Aatt/1'>AATT minor groove</scene> which has a nearly 3 angstrom narrower interphosphate distance than a typical minor groove, due to the consecutive A/T bases. This unique trait helps account for MeCP2’s exquisite precision.<ref name="Bird"/> | The crystal structure of the DNA binding domain of MeCP2 was solved by Ho et al. in 2008 providing researchers with critical new information about the cause of RTT at the molecular level. <scene name='Methyl_CpG_Binding_Protein_2/Protein/3'>The MeCP2 DNA Binding Domain (MBD)</scene> binds the <scene name='Methyl_CpG_Binding_Protein_2/Dna/3'>methylated BDNF DNA sequence</scene> using a predominantly <scene name='Methyl_CpG_Binding_Protein_2/Hydrophil/1'>hydrophilic pocket</scene>. MeCP2 recognition of the mCpG sequence involves <scene name='Methyl_CpG_Binding_Protein_2/Water/1'>five water molecules</scene> each making CH-O hydrogen bonds. <scene name='Methyl_CpG_Binding_Protein_2/Water_1/1'>W22 forms hydrogen bonds</scene> with Asp 121, W24, W21, and N4 of m5C33. <scene name='Methyl_CpG_Binding_Protein_2/Water_2/3'>W24 forms hydrogen bonds</scene> with Tyr 123, Arg 133, water 22, and both N4 of m5C8 and a CH-O interaction with the methyl group of m5C8. The only residues that directly interact with DNA bases <scene name='Methyl_CpG_Binding_Protein_2/Asp/1'>are Asp 121</scene>, forming a CH-O hydrogen bond with methyl of m5C8, <scene name='Methyl_CpG_Binding_Protein_2/Fingers/1'>Arg 111, and Arg 133</scene>, each of which form symmetrical hydrogen bonds with each guanine in the mCpG pair. Both of these “Arginine Fingers” <scene name='Methyl_CpG_Binding_Protein_2/Coplanar/2'>lie in a plane with the guanine bases</scene> and are locked into position by <scene name='Methyl_CpG_Binding_Protein_2/Salt_bridges/1'>salt bridges with Asp 121 and Glu 137</scene>, placing the gaunidinum groups directly <scene name='Methyl_CpG_Binding_Protein_2/Methylated/1'>above/below the methyl groups </scene> of the methylated cytidine bases.<ref name="Bird"/> The C-terminal region of <scene name='Methyl_CpG_Binding_Protein_2/Mbd/1'>the MBD</scene> includes an unusual <scene name='Methyl_CpG_Binding_Protein_2/Tandem/1'>tandem Asx-ST motif</scene>, that consists of an Asx turn (Residues 156-158) followed by an ST motif (158-161). The <scene name='Methyl_CpG_Binding_Protein_2/Asx/1'>Asx turn is formed</scene> by a hydrogen bond that connects the main chain nitrogen of Thr 158 and the side chain of Asp 156, while the <scene name='Methyl_CpG_Binding_Protein_2/St/2'>ST motif is held together </scene> by hydrogen bonds to Thr 158. T158M, which is the most common missense mutation causing Rett Syndrome abolishes DNA binding because it disrupts this Asx-ST motif. Another well-known mutation, causing RTT, R106W, disrupts the motif stabilizing hydrogen bonds formed between <scene name='Methyl_CpG_Binding_Protein_2/106/2'>Arg 106 and Thr 158 and Val 159</scene>. This Asx-ST motif stabilizes MeCP2’s interaction with DNA by specifically binding to the <scene name='Methyl_CpG_Binding_Protein_2/Aatt/1'>AATT minor groove</scene> which has a nearly 3 angstrom narrower interphosphate distance than a typical minor groove, due to the consecutive A/T bases. This unique trait helps account for MeCP2’s exquisite precision.<ref name="Bird"/> | ||
Revision as of 04:15, 13 March 2011
| |||||||||||
Additional Structures of MeCP2
MeCP2 is present in mature nerve cells and is involved in turning off several genes. Human MeCP2 are called MBD1 to MBD4 and contain a methyl-CpG binding domain (MBD) which binds to methylated DNA.
1ig4 – hMBD1 MBD domain + DNA – human – NMR
1d9n - hMBD1 MBD domain – NMR
3iho – hMBD4 glycosylate domain
1qk9 - hMeCP2 MBD domain - NMR
3c2i – hMeCP2 MBD domain (mutant) + DNA
1ub1 – MeCP2 attachment-region binding domain – chicken – NMR
1ngn - MBD4 glycosylate domain - mouse
References
- ↑ 1.0 1.1 1.2 1.3 Ho KL, McNae IW, Schmiedeberg L, Klose RJ, Bird AP, Walkinshaw MD. MeCP2 binding to DNA depends upon hydration at methyl-CpG. Mol Cell. 2008 Feb 29;29(4):525-31. PMID:18313390 doi:10.1016/j.molcel.2007.12.028
- ↑ Zoghbi HY. Postnatal neurodevelopmental disorders: meeting at the synapse? Science. 2003 Oct 31;302(5646):826-30. PMID:14593168 doi:10.1126/science.1089071
- ↑ Marchetto MC, Carromeu C, Acab A, Yu D, Yeo GW, Mu Y, Chen G, Gage FH, Muotri AR. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010 Nov 12;143(4):527-39. PMID:21074045 doi:10.1016/j.cell.2010.10.016
- ↑ Guy J, Gan J, Selfridge J, Cobb S, Bird A. Reversal of neurological defects in a mouse model of Rett syndrome. Science. 2007 Feb 23;315(5815):1143-7. Epub 2007 Feb 8. PMID:17289941 doi:10.1126/science.1138389
Proteopedia Page Contributors and Editors (what is this?)
David Canner, Michal Harel, Eric Martz, Jaime Prilusky, Alexander Berchansky