We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Helicase
From Proteopedia
(Difference between revisions)
| Line 29: | Line 29: | ||
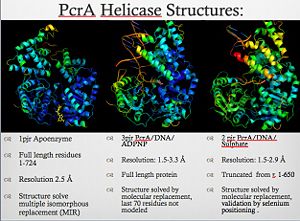
[[Image:Pcr4_structures.jpg|thumb|300px|left|PcrA_Structure]] | [[Image:Pcr4_structures.jpg|thumb|300px|left|PcrA_Structure]] | ||
| + | {{clear}} | ||
PcrA is part of the replication machinery of the [http://en.wikipedia.org/wiki/Geobacillus_stearothermophilus Geobacillus stearothermophilus]a gram (+) bacteria, This helicase is part of the superfamily I of Helicases. Monomeric protein that is mainly <scene name='User:Luis_E_Ramirez-Tapia/Sandbox_1/Initial/1'>alpha helical</scene> has the <scene name='User:Luis_E_Ramirez-Tapia/Sandbox_2/1pjrconser/2'>highly conserved</scene> Rec domians. This helicase was reported as a mutation in the gen PcrA from [http://en.wikipedia.org/wiki/staphylococcu "Stapphylococcus aerous"], this mutation was related to a deficiency in the replication of a reporter plasmid.[http://www.ncbi.nlm.nih.gov/pubmed/8232203?ordinalpos=81&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_DefaultReportPanel.Pubmed_RVDocSum] | PcrA is part of the replication machinery of the [http://en.wikipedia.org/wiki/Geobacillus_stearothermophilus Geobacillus stearothermophilus]a gram (+) bacteria, This helicase is part of the superfamily I of Helicases. Monomeric protein that is mainly <scene name='User:Luis_E_Ramirez-Tapia/Sandbox_1/Initial/1'>alpha helical</scene> has the <scene name='User:Luis_E_Ramirez-Tapia/Sandbox_2/1pjrconser/2'>highly conserved</scene> Rec domians. This helicase was reported as a mutation in the gen PcrA from [http://en.wikipedia.org/wiki/staphylococcu "Stapphylococcus aerous"], this mutation was related to a deficiency in the replication of a reporter plasmid.[http://www.ncbi.nlm.nih.gov/pubmed/8232203?ordinalpos=81&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_DefaultReportPanel.Pubmed_RVDocSum] | ||
| Line 39: | Line 40: | ||
[[Image:Wigleypcr4.jpg|thumb|300px|left]] | [[Image:Wigleypcr4.jpg|thumb|300px|left]] | ||
| - | + | {{clear}} | |
==PcrA Helicase Mechanism : The Mexican Wave== | ==PcrA Helicase Mechanism : The Mexican Wave== | ||
| - | {{STRUCTURE_1pjr| PDB=3pjr | SCENE='User:Luis_E_Ramirez-Tapia/Sandbox_2/3pjrinitial/1'}} | ||
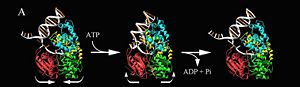
Professor Dale B. Wigley' group in 1996-1999 was able to crystalize the intermediate states from PcrA, giving solution to the controversy of what kind of mechanism this helicase has. [http://www.ncbi.nlm.nih.gov/pubmed/10199404ordinalpos=39&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_DefaultReportPanel.Pubmed_RVDocSum] | Professor Dale B. Wigley' group in 1996-1999 was able to crystalize the intermediate states from PcrA, giving solution to the controversy of what kind of mechanism this helicase has. [http://www.ncbi.nlm.nih.gov/pubmed/10199404ordinalpos=39&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_DefaultReportPanel.Pubmed_RVDocSum] | ||
Two crystal form of the enzyma, one couple with a 10 mer DNA and a non hydrolizable form of ATP (ATPnP) (pdb id: [[3pjr]], <scene name='User:Luis_E_Ramirez-Tapia/Sandbox_2/3pjrinitial/1'> (Enzyme Subtrate Structure) </scene>and another a truncated form embebed in sulfate (pdb id: [[2pjr]]<scene name='User:Luis_E_Ramirez-Tapia/Sandbox_2/2pjrinitial/1'> (Enzyme Product Structure)</scene>, give a light in a model for how ATP hydrolysis results in motor movement along ssDNA. In the figure below step 1 (top) is the ATP free (product) ssDNA conformation. The DNA bases are labelled arbitrarily. On binding ATP, F626 creates a new binding pocket for base 6. Likewise, F64 destroys an acceptor pocket for base 2, forcing it to move to the position occupied by base 1. After ATP hydrolysis, the grip on base 6 is released. When the Y257 pocket is re-opened due to movement of F64, bases 3-6 can now flip through the acceptor pockets to their new positions. This model predicts that each ATP hydrolysis event will advance PcrA one base along ssDNA.[http://www.icnet.uk/labs/wigley/projects/helicase/35.html] | Two crystal form of the enzyma, one couple with a 10 mer DNA and a non hydrolizable form of ATP (ATPnP) (pdb id: [[3pjr]], <scene name='User:Luis_E_Ramirez-Tapia/Sandbox_2/3pjrinitial/1'> (Enzyme Subtrate Structure) </scene>and another a truncated form embebed in sulfate (pdb id: [[2pjr]]<scene name='User:Luis_E_Ramirez-Tapia/Sandbox_2/2pjrinitial/1'> (Enzyme Product Structure)</scene>, give a light in a model for how ATP hydrolysis results in motor movement along ssDNA. In the figure below step 1 (top) is the ATP free (product) ssDNA conformation. The DNA bases are labelled arbitrarily. On binding ATP, F626 creates a new binding pocket for base 6. Likewise, F64 destroys an acceptor pocket for base 2, forcing it to move to the position occupied by base 1. After ATP hydrolysis, the grip on base 6 is released. When the Y257 pocket is re-opened due to movement of F64, bases 3-6 can now flip through the acceptor pockets to their new positions. This model predicts that each ATP hydrolysis event will advance PcrA one base along ssDNA.[http://www.icnet.uk/labs/wigley/projects/helicase/35.html] | ||
| Line 52: | Line 52: | ||
==The Superfamily 1 (SF1)== | ==The Superfamily 1 (SF1)== | ||
| - | PcrA share structural domains with the Rec helicases, like UvrD and RepD from E. coli, Superfamily 1 (SF1) helicases are probably the best characterized class, certainly from a structural perspective. All members characterized to date are bona fide helicases and α enzymes. Indeed, from their mode of translocation via the bases it is difficult to envisage how they could translocate along a duplex. However, they can have either A or B directional polarity. | + | PcrA share structural domains with the Rec helicases, like UvrD ([[2is1]]) and RepD ([[1uaa]]) from E. coli, Superfamily 1 (SF1) helicases are probably the best characterized class, certainly from a structural perspective. All members characterized to date are bona fide helicases and α enzymes. Indeed, from their mode of translocation via the bases it is difficult to envisage how they could translocate along a duplex. However, they can have either A or B directional polarity. |
{{clear}} | {{clear}} | ||
Revision as of 10:28, 10 April 2013
| |||||||||||
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Wayne Decatur, Joel L. Sussman