Helicase
From Proteopedia
| Line 1: | Line 1: | ||
| + | __NOTOC__ | ||
<StructureSection load='1pjr' size='450' side='right' scene='' caption=''> | <StructureSection load='1pjr' size='450' side='right' scene='' caption=''> | ||
| + | __NOTOC__ | ||
'''Helicase''' (Hel) is a motor protein which separates nucleic acid strands like DNA double helix or self-annealed RNA. They use ATP hydrolysis for energy. Hel falls into 5 superfamilies (SF1-SF5). Some Hel contain a Helicase and RNase D C terminal | '''Helicase''' (Hel) is a motor protein which separates nucleic acid strands like DNA double helix or self-annealed RNA. They use ATP hydrolysis for energy. Hel falls into 5 superfamilies (SF1-SF5). Some Hel contain a Helicase and RNase D C terminal | ||
Domain (HRDC). The α-thalassemia and mental retardation X-linked syndrome helicase (ATRX ), contains an ATRX-Dnmt3-Dnmt3L (ADD) domain in which many disease-related mutations are found. | Domain (HRDC). The α-thalassemia and mental retardation X-linked syndrome helicase (ATRX ), contains an ATRX-Dnmt3-Dnmt3L (ADD) domain in which many disease-related mutations are found. | ||
Revision as of 10:29, 10 April 2013
Helicase (Hel) is a motor protein which separates nucleic acid strands like DNA double helix or self-annealed RNA. They use ATP hydrolysis for energy. Hel falls into 5 superfamilies (SF1-SF5). Some Hel contain a Helicase and RNase D C terminal Domain (HRDC). The α-thalassemia and mental retardation X-linked syndrome helicase (ATRX ), contains an ATRX-Dnmt3-Dnmt3L (ADD) domain in which many disease-related mutations are found. For details of PcrA helicase see For ATP-dependent helicase Rho see For ATP-dependent helicase Q see For ATP-dependent helicase RecG see For DEAD box ATP-dependent RNA helicase see
See also What is a Helicase?Helicases are nucleic acid–dependent ATP-ases that are capable of unwinding DNA [1] or RNA [2] duplex substrates. As a consequence, they play roles in almost every process in cells that involves nucleic acids, including DNA replication and repair, transcription, translation, ribosome synthesis (1). PcrA a Simple Model for HelicasesPcrA is part of the replication machinery of the Geobacillus stearothermophilusa gram (+) bacteria, This helicase is part of the superfamily I of Helicases. Monomeric protein that is mainly has the Rec domians. This helicase was reported as a mutation in the gen PcrA from "Stapphylococcus aerous", this mutation was related to a deficiency in the replication of a reporter plasmid.[3]
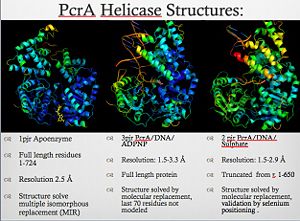
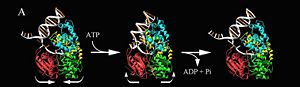
PcrA BiochemistryPcrA is has an ATPas activityt which directionality is from 3' to 5' helicase strand separation reaction. The enzyme shows a specificity for the DNA substrate in gel mobility assays with the preferred substrate being one containing both single and double stranded regions of DNA. In contrast to Rep and UvrD from E. coli, there is not evidence for dimerisation of the enzyme using gel filtration, or by crosslinking in the presence of combinations of Mg2+, nucleotides and DNA. Moreover, kcat for ATP hydrolysis is constant over a large range of protein concentrations. Therefore, the protein appears to be monomeric under all conditions tested, including in the structure of two crystal forms of PcrA.[4] PcrA Helicase Mechanism : The Mexican WaveProfessor Dale B. Wigley' group in 1996-1999 was able to crystalize the intermediate states from PcrA, giving solution to the controversy of what kind of mechanism this helicase has. [5] Two crystal form of the enzyma, one couple with a 10 mer DNA and a non hydrolizable form of ATP (ATPnP) (pdb id: 3pjr, and another a truncated form embebed in sulfate (pdb id: 2pjr, give a light in a model for how ATP hydrolysis results in motor movement along ssDNA. In the figure below step 1 (top) is the ATP free (product) ssDNA conformation. The DNA bases are labelled arbitrarily. On binding ATP, F626 creates a new binding pocket for base 6. Likewise, F64 destroys an acceptor pocket for base 2, forcing it to move to the position occupied by base 1. After ATP hydrolysis, the grip on base 6 is released. When the Y257 pocket is re-opened due to movement of F64, bases 3-6 can now flip through the acceptor pockets to their new positions. This model predicts that each ATP hydrolysis event will advance PcrA one base along ssDNA.[6]
The Superfamily 1 (SF1)PcrA share structural domains with the Rec helicases, like UvrD (2is1) and RepD (1uaa) from E. coli, Superfamily 1 (SF1) helicases are probably the best characterized class, certainly from a structural perspective. All members characterized to date are bona fide helicases and α enzymes. Indeed, from their mode of translocation via the bases it is difficult to envisage how they could translocate along a duplex. However, they can have either A or B directional polarity.
3D structures of helicaseUpdated on 10-April-2013 3oc3 – Hel MOT1 heat domain + TFIID-1 – Encephalitozoon cuniculi 2p6r – AfHel 308 + DNA – Archaeoglobus fulgidus DNA helicase3lfu – EcHel II – Escherichia coli ATP-dependent helicase ATRX3qln - hATRX ADD domain – human ATP-dependent helicase Rho3l0o – TmRho – Thermotoga maritima ATP-dependent helicase MFD3hjh – EcMFD motor domain ATP-dependent helicase E12v9p – PvE1 helicase domain – Bovine papillomavirus ATP-dependent helicase CHD12h1e, 2xb0, 3mwy – yCHD1 chromodomain Dead box ATP-dependent RNA helicase3fho – DBP5 – fission yeast Dead box ATP-dependent RNA helicase complexes3rrm – yDBP5 residues 91-482 (mutant) + GLE1 + NUP159 + ADP + IP6 – yeast ATP-dependent RNA helicase2xzl – yNAM7 CH and helicase domains + RNA Viral ATP-dependent RNA helicase NS33o8b, 3o8d, 1cu1, 8ohm, 1hei – HvHel NS3 – Hepatitis C virus Viral ATP-dependent RNA helicase NS3 complexes3o8c, 3o8r, 3kqh, 3kqk, 3kql, 3kqn, 3kqu, 1a1v - HvHel NS3 + RNA ATP-dependent RNA helicase SUV33rc3 – hSUV3 residues 47-722 ATP-dependent RNA helicase A3llm – hHel A ATP-dependent RNA helicase Repa1uaa – EcRepa + DNA ATP-dependent RNA helicase HEF1x2i – HEF DNA-binding domain – Pyrococcus furiosus Werner syndrome ATP-dependent DNA helicase2e1e, 2e1f, 2dgz – hWRN HRDC domain ATP-dependent DNA helicase Q2v1x - hQ1 residues 49-616 ATP-dependent DNA helicase RecG1gm5 – TmRecG + DNA ATP-dependent DNA helicase RecQ3iuo – RecQ residues 604-725 – Porphyromonas gingivalis ATP-dependent DNA helicase RuvA, RuvB2ztc, 2ztd, 2zte, 2h5x – MtRuvA – Mycobacterium tuberculosis ATP-dependent DNA helicase UVSW2oca, 1rif- T4UVSW (mutant) – Enterobacteria phage T4 ATP-dependent DNA helicase UVRD2is1 - EcUVRD + DNA ATP-dependent DNA helicase PriA2d7e – EcPriA DNA-binding domain ATP-dependent DNA helicase PcrA1qhg – GsPcrA (mutant) + ADPNP – Geobacillus stearothermophilus DnaB helicase3gxv – HpDnaB – Helicobacter pylori Helicase NSP22hwk – NSP2 – Venzuelan equine encephalitis virus Helicase SNF21z5z – SsSNF2 C terminal – Sulfolobus solfataricus Helicase SKI22va8 – SsSKI2 3crv, 3crw – XPD – Sulfolobus acidocaldarius 3hib, 3im1, 3im2 – yBrr2 2nd SEC63 domain ReferencesCrystal structure of a DExx box DNA helicase., Subramanya HS, Bird LE, Brannigan JA, Wigley DB, Nature. 1996 Nov 28;384(6607):379-83. PMID:8934527
| |||||||||||||
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Wayne Decatur, Joel L. Sussman