We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
SUMO
From Proteopedia
(Difference between revisions)
| Line 15: | Line 15: | ||
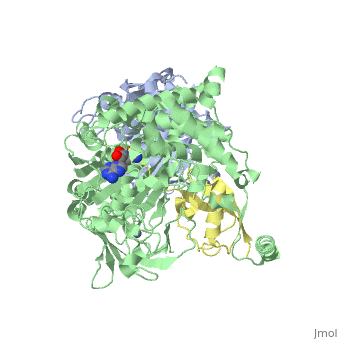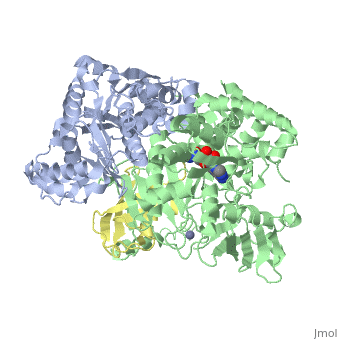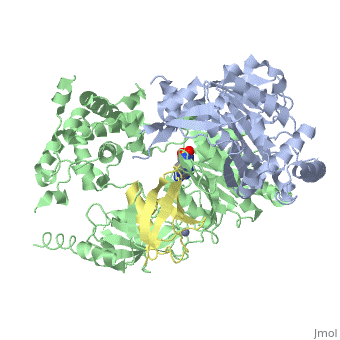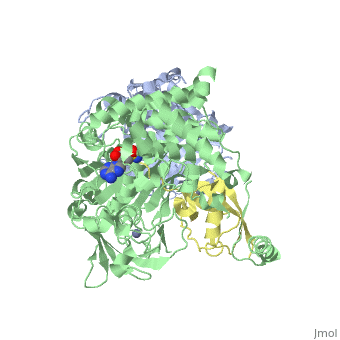
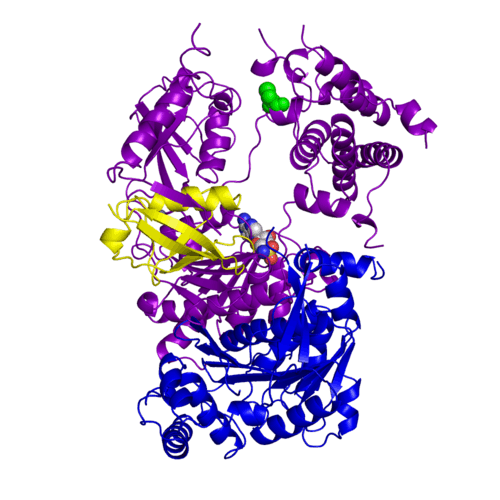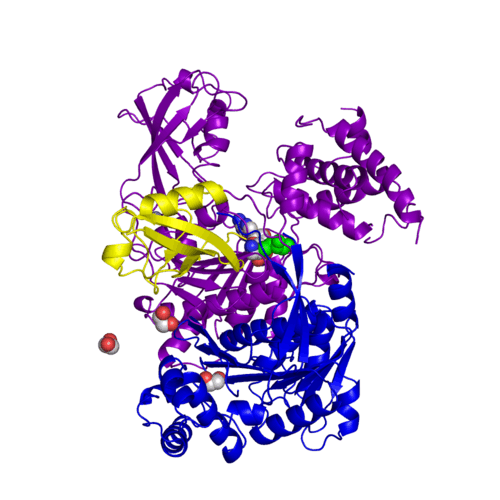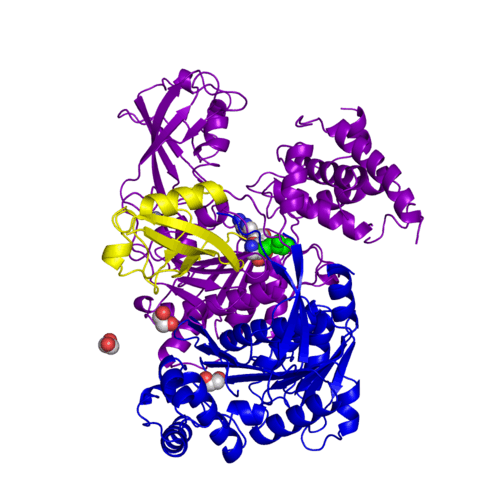
The <scene name='3kyc/Al/2'>structural alignment</scene> of the crystal structures for human SUMO E1 in complex with SUMO adenylate (AMSN) and tetrahedral intermediate (AVSN) analogues revealed opened conformation (<font color='orange'><b>SUMO1 in orange</b></font>, <font color='blue'><b>SAE1 colored in blue</b></font>, and <font color='darkviolet'><b>other domains in darkviolet</b></font>) and closed conformation (<font color='yellow'><b>SUMO1 in yellow</b></font>, <font color='cyan'><b>SAE1 colored in cyan</b></font>, and <font color='magenta'><b>other domains in magenta</b></font>), respectively. In the <scene name='3kyc/Al/7'>open conformation</scene> ([[3kyc]]) the distance between Cys domain (including Cys173) and mimic of the acyl adenylate intermediate AMSN is very long, while in the <scene name='3kyc/Al/6'>closed conformation</scene> ([[3kyd]]), the catalytic Cys173 is posioned near AVSN and SUMO1, so the overall structure revealed dramatic rearrangement. This large conformational change forms the <scene name='3kyc/Al/8'>E1~SUMO1-AVSN tetrahedral intermediate analogue</scene>.<ref>PMID:20164921</ref> | The <scene name='3kyc/Al/2'>structural alignment</scene> of the crystal structures for human SUMO E1 in complex with SUMO adenylate (AMSN) and tetrahedral intermediate (AVSN) analogues revealed opened conformation (<font color='orange'><b>SUMO1 in orange</b></font>, <font color='blue'><b>SAE1 colored in blue</b></font>, and <font color='darkviolet'><b>other domains in darkviolet</b></font>) and closed conformation (<font color='yellow'><b>SUMO1 in yellow</b></font>, <font color='cyan'><b>SAE1 colored in cyan</b></font>, and <font color='magenta'><b>other domains in magenta</b></font>), respectively. In the <scene name='3kyc/Al/7'>open conformation</scene> ([[3kyc]]) the distance between Cys domain (including Cys173) and mimic of the acyl adenylate intermediate AMSN is very long, while in the <scene name='3kyc/Al/6'>closed conformation</scene> ([[3kyd]]), the catalytic Cys173 is posioned near AVSN and SUMO1, so the overall structure revealed dramatic rearrangement. This large conformational change forms the <scene name='3kyc/Al/8'>E1~SUMO1-AVSN tetrahedral intermediate analogue</scene>.<ref>PMID:20164921</ref> | ||
| + | |||
| + | [[Image:kyc.gif|left|500px]] | ||
| + | For better understanding of the difference between these two conformations you can see this [[Morphs|morph]] (generated by using [http://polyview.cchmc.org/polyview3d.html POLYVIEW-3D: http://polyview.cchmc.org/polyview3d.html]; reload/refresh this page to restart this movie). Of note, in contrast to the previous figure, the same domains of these two structures ([[3kyc]] and [[3kyd]]) are colored in the same colors (<font color='yellow'><b>SUMO1 in yellow</b></font>, <font color='blue'><b>SAE1 colored in blue</b></font> and <font color='darkviolet'><b>other domains in darkviolet</b></font>). The catalytic Cys173 is shown in the spacefill representation and colored green, AMSN (or AVSN) are shown in the spacefill representation and colored in CPK colors. | ||
</StructureSection> | </StructureSection> | ||
{{Clear}} | {{Clear}} | ||
| - | |||
| - | [[Image:kyc.gif|left|350px]] | ||
| - | For better understanding of the difference between these two conformations you can see this [[Morphs|morph]] (generated by using [http://polyview.cchmc.org/polyview3d.html POLYVIEW-3D: http://polyview.cchmc.org/polyview3d.html]; reload/refresh this page to restart this movie). Of note, in contrast to the previous figure, the same domains of these two structures ([[3kyc]] and [[3kyd]]) are colored in the same colors (<font color='yellow'><b>SUMO1 in yellow</b></font>, <font color='blue'><b>SAE1 colored in blue</b></font> and <font color='darkviolet'><b>other domains in darkviolet</b></font>). The catalytic Cys173 is shown in the spacefill representation and colored green, AMSN (or AVSN) are shown in the spacefill representation and colored in CPK colors. | ||
| - | {{Clear}} | ||
| - | |||
== 3D Structures of SUMO == | == 3D Structures of SUMO == | ||
Revision as of 10:42, 7 November 2017
| |||||||||||
3D Structures of SUMO
Updated on 07-November-2017
Reference
- ↑ Sarge KD, Park-Sarge OK. Sumoylation and human disease pathogenesis. Trends Biochem Sci. 2009 Apr;34(4):200-5. doi: 10.1016/j.tibs.2009.01.004. Epub, 2009 Mar 11. PMID:19282183 doi:http://dx.doi.org/10.1016/j.tibs.2009.01.004
- ↑ Olsen SK, Capili AD, Lu X, Tan DS, Lima CD. Active site remodelling accompanies thioester bond formation in the SUMO E1. Nature. 2010 Feb 18;463(7283):906-12. PMID:20164921 doi:10.1038/nature08765