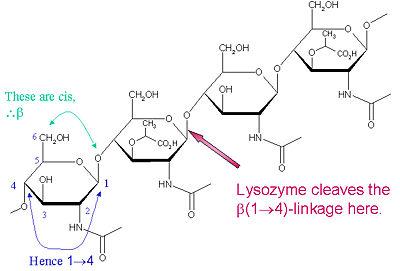
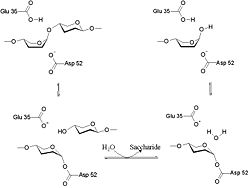
Hen Egg-White (HEW) Lysozyme
From Proteopedia
| Line 1: | Line 1: | ||
| - | + | <StructureSection load='1hew' size='450' side='right' scene='' caption=''>__NOTOC__ | |
| - | + | ||
==Introduction== | ==Introduction== | ||
| Line 9: | Line 8: | ||
The characterization of lysozyme in 1922 by Alexander Fleming was providential in that the undertaken experiment related to the discovery of lysozyme was not geared toward any knowledge of such a protein as lysozyme <ref>Lysozyme. 2008. Lysozyme.co.uk. http://lysozyme.co.uk/</ref>. During the unrelated experiment, nasal drippings were inadvertently introduced to a petri dish containing a bacterial culture, which culture consequently exhibited the results of an as yet unknown enzymatic reaction. The observation of this unknown reaction led to further research on the components of this reaction as well as to the corresponding identification of the newfound "lysozyme." Fleming's discovery was complemented by David C. Phillips' 1965 description of the three-dimensional structure of lysozyme via a 200 pm resolution model obtained from X-ray crystallography <ref>Lysozyme, 2008. Lysozyme.co.uk. http://lysozyme.co.uk/</ref>. Phillips' work was especially groundbreaking since Phillips had managed to successfully elucidate the structure of an enzyme via X-ray crystallography - a feat that had never before been accomplished<ref>Bugg, T. 1997. An Introduction to Enzyme and Coenzyme Chemistry. Blackwell Science Ltd., Oxford </ref>. Phillips' research also led to the first sufficiently described enzymatic mechanism of catalytic action <ref>1967. Proc R Soc Lond B Bio 167 (1009): 389–401.</ref>. Thus, Phillips' elucidation of the function of lysozyme led Phillips to reach a more general conclusion on the diversity of enzymatic chemical action in relation to enzymatic structure. Clearly, the findings of Phillips as well as the more general historical development of the understanding of the structure and function of lysozyme have been paramount to the more general realm of enzyme chemistry. See also [[Molecular Playground/Lysozyme]] and [[User:Judy Voet/Lysozyme]]. | The characterization of lysozyme in 1922 by Alexander Fleming was providential in that the undertaken experiment related to the discovery of lysozyme was not geared toward any knowledge of such a protein as lysozyme <ref>Lysozyme. 2008. Lysozyme.co.uk. http://lysozyme.co.uk/</ref>. During the unrelated experiment, nasal drippings were inadvertently introduced to a petri dish containing a bacterial culture, which culture consequently exhibited the results of an as yet unknown enzymatic reaction. The observation of this unknown reaction led to further research on the components of this reaction as well as to the corresponding identification of the newfound "lysozyme." Fleming's discovery was complemented by David C. Phillips' 1965 description of the three-dimensional structure of lysozyme via a 200 pm resolution model obtained from X-ray crystallography <ref>Lysozyme, 2008. Lysozyme.co.uk. http://lysozyme.co.uk/</ref>. Phillips' work was especially groundbreaking since Phillips had managed to successfully elucidate the structure of an enzyme via X-ray crystallography - a feat that had never before been accomplished<ref>Bugg, T. 1997. An Introduction to Enzyme and Coenzyme Chemistry. Blackwell Science Ltd., Oxford </ref>. Phillips' research also led to the first sufficiently described enzymatic mechanism of catalytic action <ref>1967. Proc R Soc Lond B Bio 167 (1009): 389–401.</ref>. Thus, Phillips' elucidation of the function of lysozyme led Phillips to reach a more general conclusion on the diversity of enzymatic chemical action in relation to enzymatic structure. Clearly, the findings of Phillips as well as the more general historical development of the understanding of the structure and function of lysozyme have been paramount to the more general realm of enzyme chemistry. See also [[Molecular Playground/Lysozyme]] and [[User:Judy Voet/Lysozyme]]. | ||
| - | + | ||
==Function== | ==Function== | ||
Revision as of 10:18, 8 May 2013
| |||||||||||
Contents |
3D Structures of Lysozyme
Updated February 2013
Lys
2x0a, 3iju, 3ijv, 3a8z, 2w1y, 2w1m, 2w1x, 2w1l, 3e3d, 3exd, 2zq3, 2zq4, 3b72, 3b6l, 2z12, 2z18, 2z19, 2vb1, 2hu3, 2hub, 2htx, 2hu1, 2yvb, 2epe, 2g4p, 2g4q, 2cgi, 2b5z, 2d4k, 2d91, 2f2n, 2fbb, 2c8o, 2c8p, 2a6u, 2aub, 2blx, 2bly, 2a7d, 2a7f, 1wtm, 1wtn, 1w6z, 1vdp, 1vdq, 1vds, 1vdt, 1ved, 1v7t, 1ps5, 2cds, 1lj3, 1lj4, 1lje, 1ljf, 1ljg, 1ljh, 1lji, 1ljj, 1ljk, 1jis, 1jit, 1jiy, 1jj0, 1jj1, 1jj3, 1iee, 1qio, 1f0w, 1f10, 1dpx, 1c10, 1qtk, 1lz8, 1lz9, 1bhz, 1bgi, 1bwh, 1bwi, 1bwj, 1bvx, 1hsw, 1hsx, 1lpi, 4lzt, 3lzt, 1aki, 1jpo, 1rfp, 193l, 194l, 5lym, 1lza, 3lyt, 4lyt, 5lyt, 6lyt, 2lzt, 1lzt, 1lzh, 2lzh, 7lyz, 1lyz, 2lyz, 3lyz, 4lyz, 5lyz, 6lyz, 1lsg, 1hc0, 1vau, 3agg, 3agh, 3agi, 3ajn, 3aw6, 3aw7, 3p4z, 3p64, 3p65, 3p66, 3p68, 3qy4, 3rnx, 3rw8, 3rz4, 3n9a, 3n9c, 3n9e, 3rt5, 3sp3, 3t6u, 3atn, 4dd0, 4dd1, 4dd2, 4dd3, 3ru5, 3tmu, 3tmv, 3tmw, 3tmx, 3zek, 4aga, 4axt, 4b0d, 4b49, 4b4e, 4b4i, 4b4j, 4e3u, 4et8, 4et9, 4eta, 4etb, 4etc, 4etd, 4ete, 4g49, 4g4a, 4g4b, 4g4c, 4g4h, 4gcb, 4gcc, 4gcd, 4gce, 4gcf, 4h1p - cHEWL – chicken
3zvq – cHEWL proteolyzed
3ar3, 3ok0 – cHEWL (mutant)br />
1xei, 1xej, 1xek, 1uco, 1lma, 4lym – HEWL low hydration
1lsa, 1lsb, 1lsc, 1lsd, 1lse, 1lsf, 1lys – HEWL temperature influence
2lym, 3lym – HEWL pressure influence
132l – HEWL methylated
1rcm – HEWL 3 S-S form
2xbr, 2xbs- HEWL– Raman crystallography
2zwb, 1io5, 1lzn - HEWL– Neutron
1gxv, 1gxx, 1e8l - HEWL- NMR
2hs7, 2hso, 2hs9- HEWL– Powder diffraction
1ir7, 1ir8, 1ir9, 1ioq, 1ior, 1ios, 1iot, 1flq, 1flu, 1flw, 1fly, 1fn5, 1kxw, 1kxx, 1kxy, 1uia, 1uib, 1uic, 1uid, 1uie, 1uif, 1uig, 1uih, 1lsm, 1lsn, 1hel, 1hem, 1hen, 1heo, 1hep, 1heq, 1her – HEWL (mutant)
1lyo, 2lyo, 3lyo, 4lyo – HEWL cross-linked
1xft - tuLys – turkey - Powder diffraction
1jse, 1tew, 135l, 2lz2, 1lz2 – tuLys
3lz2 – tuLys - Laue
3ab6 – Lys + NAG3 – Hard clam
2x8r – Lys GH25 – Aspergillus fumigatus
3mgw – Lys G – Atlantic salmon
3gxk, 3gxr – Lys G + NAG – Atlantic cod
2fbd – HfLys 1
3cb7 – HfLys 2
2zij, 2zik, 2zil, 2nwd, 1iwt, 1iwu, 1iwv, 1iww, 1iwx, 1iwy, 1iwz, 1jwr, 1jsf, 1rex, 1lz1 – hLys – human
1w08, 1ix0, 1ioc, 1ip1, 1ip2, 1ip3, 1ip4, 1ip5, 1ip6, 1ip7, 1qsw, 1gev, 1gez, 1gf0, 1gf3, 1gf4, 1gf5, 1gf6, 1gf7, 1i1z, 1i20, 1i22, 1gfr, 1gft, 1gfu, 1gfv, 1gf8, 1gf9, 1gfa, 1gfe, 1gfg, 1gfh, 1gfj, 1gfk, 1inu, 1gdx, 1ge0, 1ge1, 1ge2, 1ge3, 1ge4, 1gdw, 1gaz, 1gb0, 1gb2, 1gb3, 1gb5, 1gb6, 1gb7, 1gb8, 1gb9, 1gbo, 1gbw, 1gbx, 1gby, 1gbz, 1gay, 1eq4, 1eq5, 1eqe, 1c7p, 1di3, 1di4, 1di5, 1c43, 1c45, 1c46, 1ckg, 1cj6, 1cj7, 1cj8, 1cj9, 1ckc, 1ckd, 1ckf, 1ckh, 1b5z, 1b70, 1b7q, 1b7r, 1b7s, 1b7l, 1b7m, 1b7n, 1b7p, 1b5u, 1b5v, 1b5w, 1b5x, 1b5y, 1bb3, 1bb4, 2bqa, 2bqb, 2bqc, 2bqd, 2bqe, 2bqf, 2bqg, 2bqh, 2bqi, 2bqj, 2bqk, 2bql, 2bqm, 2bqn, 2bqo, 2mea, 2meb, 2mec, 2med, 2mee, 2mef, 2meg, 2meh, 2mei, 1wqm, 1wqn, 1wqo, 1wqp, 1wqq, 1wqr, 2heb, 2hea, 2hec, 2hed, 2hee, 2hef, 1jka, 1jkb, 1jkc, 1jkd, 1loz, 1lyy, 1oua, 1oub, 1ouc, 1oud, 1oue, 1ouf, 1oug, 1ouh, 1oui, 1ouj, 207l, 208l , 1yam, 1yan, 1yao, 1yap, 1yaq, 1lmt, 1lhh, 1lhi, 1lhj, 1lhk, 1lhl, 133l, 134l, 1lz4, 1lz5, 1lz6, 1laa, 1tay, 1tby, 1tcy, 1tdy, 2lhm, 3lhm, 1b7o, 3ln2, 3a3r, 3ojp - hLys (mutant)
1iy3, 1iy4 – hLys - NMR
2z2f – Lys – Bovine
2cwi, 1el1, 1qqy – dLys - dog
2z2e – dLys (mutant)
1i56 – dLys – NMR
2dqa – Lys – Tapes japonica
2gv0 – Lys – Soft-shelled turtle
1ivm – mLys M – NMR – mouse
1jfx – Lys – Streptomyces coelicolor
1gd6 – BmLys – Bombyx mori
2rsc – BmLys - NMR
1jug – Lys – Echidna
1dkj – BqLys – Bobwhite quail
2ihl – Lys – Japanese quail
1gbs – BsLys – Black swan
1lmn – RtLys – Rainbow trout
2eql – Lys – horse
1ghl – phLys – pheasant
1hhl – GfLys – Guinea fowl
1lhm – Lys (mutant) – yeast
1iiz – Lys – Antheraea mylitta
Lys small molecules complexes
3fe0, 2d4i, 2d4j, 1v7s - HEWL+ D2O
3m3u – HEWL Trp fluorescence
2xjw - HEWL + CO
1hf4, 1lks, 2ybh, 2ybi, 2ybj, 2ybl, 2ybm, 2ybn – HEWL + NO3
3a34, 3ems - HEWL + arginine
2xth – cLys + inhibitor K2PtBr6
3a90, 3a91, 3a92, 3a93, 3a94, 3a95, 3a96, 3kam, 2pc2, 2bpu, 1t3p, 1h87, 3qe8, 3qng – HEWL + rare earth
2d6b, 2hg0, 1vat, 1gwd, 1b2k, 1lkr, 1azf, 8lyz- HEWL+ halogen
1vat - HEWL + Xe
3p65, 1n4f, 3az4, 3az5, 3az6, 3az7, 4dd4, 4dd6, 4dd7, 4dd9, 4ddb, 4ddc, 4eni, 4dt3 – HEWL + metal
2i6z - HEWL + Pt drug
2zyp - HEWL + poly (allyl amine)
2f30, 2f4a – HEWL + urea derivative
2f4g, 1ykx, 1yky, 1ykz, 1yl0, 1yl1, 1z55 - HEWL + alcohol
2ydg – HEWL + ascorbate
1dpw – HEWL + MPD
1lcn – HEWL + SCN
2zxs - HEWL with glycine-amide
2h9j, 2h9k, 1yik, 1yil - HEWL + cyclam derivative
1b0d – HEWL + p-toluene-sulfonate
2q0m - HEWL + tricarbonylmanganese
3ato – HEWL + ethyl glycinate
4bad, 4baf, 4bap – HEWL + europium dipicolinate
2war – HEWL (mutant) + chitopentaose
2h5z – HfLys 1 + chitotetraose – House fly
1hnl – hLys + glutathione
1dkk – BqLys + NO3
3ayq – hcLys + inhibitor – Hard clam
Lys complex with glucoside
4dda - HEWL + NAG
3a3q – HEWL (mutant) + NAG
1sf4, 1sf6, 1sf7, 1sfb, 1sfg, 1ja2, 1ja4, 1ja6, 1ja7 – HEWL + NAG oligosaccharide – Powder diffraction
3ab6 – hcLys + NAG3
1ubz, 1d6p, 1d6q, 1bb5 – hLys (mutant) + glycoside
1uc0, 1re2, 1rem, 1rey, 1rez, 1lzr, 1lzs - hLys + glycoside
1ljn, 1jef, 1lzy – tuLys + glycoside
1h6m, 1at5, 1at6, 1lzb, 1lzc, 1lzd, 1lze, 1lzg, 1hew – HEWL + glycoside
1lsy, 1lsz - HEWL (mutant) + glycoside
1bb6, 1bb7 – RtLys + glycoside
1lmc – RtLys + bulgecin
1lmo, 1lmp, 1lmq – RtLys + glucoside
1lsp – BsLys + bulgecin
153l, 154l – Lys +glucoside – goose
Phage Lys
2oe4, 2oe7, 2oe9, 2oea, 1swz, 1cx6, 1qtc, 1qtd, 1qth, 1qsb, 1qs5, 1qs9, 1qtb, 256l, 206l, 167l, 168l, 169l, 170l, 171l, 172l, 173l, 174l, 175l, 176l, 177l, 178l, 181l, 182l, 183l, 184l, 185l, 186l, 187l, 188l, 1nhb, 137l, 216l, 152l, 149l, 150l, 151l, 1lyd, 2lzm, 139l, 1pdl - T4Lys – Enterobacteria phage T4
138l – T4Lys cross-linked
4lzm, 5lzm, 6lzm, 7lzm – T4Lys ionic strength
3l2x, 3k2r, 3g3v, 3g3w, 3g3x, 2q9d, 2q9e, 2igc, 2ntg, 2nth, 2ou8, 2ou9, 1zur, 1zwn, 1zyt, 2cuu, 2a4t – T4Lys (mutant) spin labeled
3l64, 3hwl, 3jr6, 3gui, 3c7w, 3c7y, 3c7z, 3c80, 3c81, 3c82, 3c83, 3c8q, 3c8r, 3c8s, 3cdo, 3cdq, 3cdr, 3cdt, 3cdv, 3f8v, 3f9l, 3fa0, 3fad, 3fi5, 3dke, 3dmv, 2o4w, 2o79, 2o7a, 2huk, 2hul, 2hum, 2b7x, 2b6t, 2b6w, 2b6x, 2b6y, 2b6z, 2b70, 2b72, 2b73, 2b74, 2b75, 1sx7, 1swy, 1sx2, 1t6h, 1ssw, 1ssy, 1t8f, 1t8g, 1t8a, 1t97, 1p56, 1p5c, 1p2l, 1p2r, 1p36, 1p37, 1p3n, 1p46, 1p64, 1p6y, 1p7s, 1pqd, 1pqi, 1pqj, 1pqk, 1pqm, 1pqo, 1oyu, 1ks3, 1kw5, 1kw7, 1ky0, 1ky1, 1l0j, 1l0k, 1lw9, 1lwg, 1lwk, 1lpy, 1llh, 1lgu, 1li6, 1jtm, 1jtn, 1jqu, 1kni, 1g06, 1g07, 1g0g, 1g0j, 1g0k, 1g0l, 1g0m, 1g0p, 1g0q, 1g1v, 1g1w, 1i6s, 257l, 258l, 260l, 1epy, 1b6i, 1cu6, 1d9w, 1ctw, 1cu0, 1cu2, 1cu3, 1cu5, 1cv1, 1cv4, 1cv5, 1cv6, 1cvk, 1cx7, 1d2w, 1d2y, 1d3f, 1d3j, 1d3m, 1d3n, 1cv3, 1qt3, 1qt4, 1qt5, 1qt6, 1qt7, 1qt8, 1qtv, 1qtz, 1qud, 1qug, 1quh, 1quo, 1qsq, 261l, 262l, 259l, 220l, 222l, 223l, 225l, 226l, 227l, 228l, 229l, 235l, 236l, 237l, 238l, 239l, 240l, 241l, 242l, 243l, 244l, 245l, 246l, 247l, 248l, 249l, 250l, 251l, 252l, 253l, 254l, 255l, 230l, 231l, 232l, 233l, 234l, 209l, 210l, 211l, 212l, 213l, 214l, 215l, 218l, 219l, 180l, 195l, 196l, 197l, 198l, 199l, 200l, 190l, 191l, 192l, 189l, 155l, 156l, 157l, 158l, 159l, 160l, 161l, 162l, 163l, 164l, 165l, 166l, 129l, 130l, 131l, 140l, 141l, 142l, 143l, 144l, 145l, 146l, 147l, 201l, 205l, 221l, 224l, 102l, 103l, 104l, 107l, 108l, 109l, 110l, 111l, 112l, 113l, 114l, 115l, 118l, 119l, 120l, 122l, 123l, 125l, 126l, 127l, 128l, 1dya, 1dyb, 1dyc, 1dyd, 1dye, 1dyf, 1dyg, 1l00, 1l85, 1l86, 1l87, 1l88, 1l89, 1l90, 1l91, 1l92, 1l93, 1l94, 1l95, 1l96, 1l97, 1l98, 1l99, 1lye, 1lyf, 1lyg, 1lyh, 1lyi, 1lyj, 217l, 1tla, 1l77, 1l79, 1l80, 1l81, 1l82, 2l78, 1l36, 1l37, 1l38, 1l39, 1l40, 1l41, 1l42, 1l43, 1l44, 1l45, 1l46, 1l47, 1l48, 1l49, 1l50, 1l51, 1l52, 1l53, 1l54, 1l55, 1l56, 1l57, 1l58, 1l59, 1l60, 1l61, 1l62, 1l63, 1l64, 1l65, 1l66, 1l67, 1l68, 1l69, 1l70, 1l71, 1l72, 1l73, 1l74, 1l75, 1l76, 1l17, 1l18, 1l19, 1l20, 1l21, 1l22, 1l23, 1l24, 1l25, 1l26, 1l27, 1l28, 1l29, 1l30, 1l31, 1l32, 1l33, 1l34, 1l35, 3lzm, 1l01, 1l02, 1l03, 1l04, 1l05, 1l06, 1l07, 1l08, 1l09, 1l10, 1l11, 1l12, 1l13, 1l14, 1l15, 1l16, 1cup, 1cuq, 1cv0, 2lc9, 2lcb, 3sb5, 3sb6, 3sb7, 3sb8, 3sb9, 3sbb, 4e97, 4ekp - T4Lys (mutant)
3sba - T4Lys (mutant) + Zn
1c60, 1c61, 1c62, 1c63, 1c64, 1c65, 1c66, 1c67, 1c68, 1c69, 1c6a, 1c6b, 1c6c, 1c6d, 1c6e, 1c6f, 1c6g, 1c6h, 1c6i, 1c6j, 1c6k, 1c6l, 1c6m, 1c6n, 1c6p, 1c6q, 1c6t - T4Lys (mutant) + noble gas
3ht6, 3ht7, 3ht8, 3ht9, 3htb, 3hu8, 3huq, 3guj, 3guk, 3gul, 3gum, 3gun, 3guo, 3gup, 3dmx, 3dmz, 3dn0, 3dn1, 3dn2, 3dn3, 3dn4, 3dn6, 3dn8, 3dna, 2rb1, 2ray, 2raz, 2rb0, 2rb2, 2rbn, 2rbo, 2rbq, 2rbr, 2rbs, 2oty,2otz, 1owy, 1owz, 1ov5, 1ov7, 1ovh, 1ovj, 1ovk, 1lgw, 1lgx, 1li2, 1li3, 1l83, 1l84, 4ekq, 4ekr, 4eks – T4Lys (mutant) + benzene derivative
3htd, 3htf, 3htg, 3hu9, 3hua, 3huk, 3hh3, 3hh4, 3hh5, 3hh6, 2rbp, 2ou0, 2f2q, 2f32, 2f47, 1xep – T4Lys (mutant) + inhibitor
3run - T4Lys (mutant) + vancomycin
148l - T4Lys (mutant) + glucoside
2z6b - T4Lys (mutant) + protein GP27
3d3d, 1d9u – lamLys + chitohexasaccharide
1am7 – lamLys - Enterobacteria phage λ
3hde, 3hdf - Lys - Enterobacteria phage p21
2anv, 2anx – Lys (mutant) - Enterobacteria phage p22
1xjt, 1xju - Lys - Enterobacteria phage p1
1lba - T7Lys - Enterobacteria phage T7
1h09, 1oba – CP1Lys - Enterobacteria phage CP-1
2ixu, 2ixv – CP1Lys + peptidoglycan analog
2j8f, 2j8g - CP1Lys (mutant) + peptidoglycan analog
1iba - T7Lys - Enterobacteria phage T7
Lys protein complex
3m18, 3g3a, 3g3b - HEWL + Variable lymphocyte receptor – Marine lamprey
3a67, 3a6b, 3a6c, 2yss, 2eiz, 2dqc, 2dqd, 2dqe, 2dqf, 2dqg, 2dqh, 2dqi, 2dqj, 1j1o, 1j1p, 1j1x, 1ic4, 1ic5, 1ic7 – HEWL + mLys antibody HYHEL-10 (mutant)
1xgp, 1xgq, 1xgr, 1xgt, 1xgu – HEWL + mLys antibody HYHEL-63
3d9a, 2eks, 1ua6, 1c08, 3hfm – HEWL + mLys antibody HYHEL-10
1uac - tuLys C + mLys antibody HYHEL-10
1yqv, 2iff – HEWL + mLys antibody HYHEL-5
1bql – BqLys + mLys antibody HYHEL-5
1ndg – HEWL + mLys antibody HYHEL-8
1ndm – HEWL + mLys antibody HYHEL-26
1dqj - HEWL + mLys antibody HYHEL-63
1nby, 1nbz – HEWL (mutant) + mLys antibody HYHEL-63
1g7h, 1g7i, 1g7j, 1g7l, 1g7m, 1kip, 1kiq, 1kir – HEWL + mAnti-HEWL monoclonal antibody (mutant)
1mlc, 1vfb - HEWL + mIGG1-κ D44.1 FAB
1a2y - HEWL (mutant) + mIGG1-κ D1.3 FV
1fdl - HEWL + mIGG1-κ D1.3 FAB
1jhl – phLys + mIGG1-κ D11.15 FV
1fbi – GfLys + mIGG1 F9.13.7 FAB
2znw, 2znx – HEWL + hSCFV10 antibody
1bvk - HEWL + hAnti Lys FV antibody
1dzb – tuLys + mSCFV 1F9
1zv5, 1zvh, 1zvy, 1zmy, 1ri8, 1rjc, 1xfp, 1jtp, 1jtt, 1jto, 1mel - HEWL + cAntibody heavy chain domain – camel
1op9 - hLys C + cAntibody heavy chain domain
3otp – HEWL + EcProtease DO – Escherichia coli
3f6z - HEWL + MLIC – Pseudomonas aeruginosa
3eba – hLys C + hCABHUL6
2qb0, 2qar – T4Lys/E80 TELSAM domain
2i25, 2i26 - HEWL +NsAntigen receptor PBLA8 variable domain – Nurse shark
1sq2, 1t6v – HEWL +NsNew Antigen receptor variable domain
1gpq – HEWL + EcInhibitor of Vertebrate Lys
4gbr – T4Lys + β-2 adrenergic receptor
1aro – T7Lys + T7 RNA polymerase
3ulr – HEWL + Src substrate cortactin + Abelson tyrosine protein kinase
4a8a, 4a8b – HEWL + protease DO
Lys chimeras
3pds - T4Lys/β-2 andrenergic receptor
3d4s – T4Lys/β-2 andrenergic receptor + cholesterol
3ny8, 3ny9 - T4Lys/β-2 andrenergic receptor + agonist
3nya - T4Lys/β-2 andrenergic receptor + antagonist
3p0g - T4Lys/β-2 andrenergic receptor + camelid antibody peptide
3sn6 - T4Lys/β-2 andrenergic receptor + camelid antibody peptide + guanine nucleotide-binding protein
3eml – T4Lys/adenosine A2a receptor + antagonist
3qak - T4Lys/adenosine A2a receptor + agonist
3odu, 3oe0, 3oe6, 3oe8, 3oe9 - T4Lys/CXCR4 chemokine receptor + antagonist
3pbl - T4Lys/dopamine receptor + antagonist
3rze - T4Lys/histamine H1 receptor + doxepin
4arj, 4epi, 4exm – T4Lys/pesticin
Some Useful External Links
Retaining Glycoside Hydrolases
References
- ↑ Lysozyme. 2010. Citizendium.org. http://en.citizendium.org/wiki/Lysozyme
- ↑ Lysozyme. 2008. Lysozyme.co.uk. http://lysozyme.co.uk/
- ↑ Lysozyme, 2008. Lysozyme.co.uk. http://lysozyme.co.uk/
- ↑ Bugg, T. 1997. An Introduction to Enzyme and Coenzyme Chemistry. Blackwell Science Ltd., Oxford
- ↑ 1967. Proc R Soc Lond B Bio 167 (1009): 389–401.
- ↑ Image from: http://www.vuw.ac.nz/staff/paul_teesdale-spittle/essentials/chapter-6/proteins/lysozyme.htm
- ↑ http://mcdb-webarchive.mcdb.ucsb.edu/sears/biochemistry/tw-enz/lysozyme/HEWL/lysozyme-overview.htm
- ↑ http://www.worthington-biochem.com/ly/default.html
- ↑ Image from: http://www.google.com/imgres?imgurl=http://www.vuw.ac.nz/staff/paul_teesdale-spittle/essentials/chapter-6/pics-and-strucs/lysozyme-mech.gif&imgrefurl=http://www.vuw.ac.nz/staff/paul_teesdale-spittle/essentials/chapter-6/proteins/lysozyme.htm&usg=__ormapG4XKg-tR5GrMSOdSMTV4vE=&h=603&w=801&sz=7&hl=en&start=17&zoom=1&tbnid=nvr9gvFrUILDkM:&tbnh=143&tbnw=189&prev=/images%3Fq%3DThe%2Blysozyme%2Breaction%2Bmechanism%26um%3D1%26hl%3Den%26sa%3DN%26biw%3D1280%26bih%3D647%26tbs%3Disch:10%2C304&um=1&itbs=1&iact=hc&vpx=521&vpy=349&dur=448&hovh=191&hovw=254&tx=140&ty=48&ei=JQ_LTPKzLIjCsAPkzt2KDg&oei=IA_LTP74OsG78gapm-GFAQ&esq=2&page=2&ndsp=18&ved=1t:429,r:2,s:17&biw=1280&bih=647
- ↑ Lysozyme, 2008. Lysozyme.co.uk. http://lysozyme.co.uk/
- ↑ Pratt, C.W., Voet, D., Voet, J.G. Fundamentals of Biochemistry - Life at the Molecular Level - Third Edition. Voet, Voet and Pratt, 2008.
- ↑ Johnson LN, Phillips DC. Structure of some crystalline lysozyme-inhibitor complexes determined by X-ray analysis at 6 Angstrom resolution. Nature. 1965 May 22;206(986):761-3. PMID:5840126
- ↑ Phillips, D. C. The hen egg white lysozyme molecule. Proc. Natl Acad. Sci. USA 57, 483-495 (1967)
- ↑ coordinates of the model kindly provided by Louise Johnson
- ↑ Vocadlo DJ, Davies GJ, Laine R, Withers SG. Catalysis by hen egg-white lysozyme proceeds via a covalent intermediate. Nature. 2001 Aug 23;412(6849):835-8. PMID:11518970 doi:10.1038/35090602
Proteopedia Page Contributors and Editors (what is this?)
Karsten Theis, Michal Harel, Anne Goodling, Alexander Berchansky, Eric Martz, Joel L. Sussman, Karl Oberholser, Jaime Prilusky, John Ripollone, Daniel Kreider, Judy Voet, John S. de Banzie