This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
SUMO
From Proteopedia
| Line 1: | Line 1: | ||
| + | <StructureSection load='3kyc' size='500' frame='true' align='right' scene='3kyc/Cv/1' > | ||
[[Image:1wm2.png|left|200px|thumb|Crystal Structure of human SUMO-2 protein, [[1wm2]]]] | [[Image:1wm2.png|left|200px|thumb|Crystal Structure of human SUMO-2 protein, [[1wm2]]]] | ||
| - | {{STRUCTURE_2pe6| PDB=2pe6 | SIZE=400| SCENE= |right|CAPTION=Human SUMO-1 protein (green) complex with Ubc9 (grey), [[2pe6]] }} | ||
| Line 22: | Line 22: | ||
| - | [[SUMO]] is a small ubiquitin-like modifier which covalently attaches to cellular proteins to modify their function. SUMO is similar in structure but not in sequence to [[Ubiquitin|ubiquitin]]. In several organisms SUMO is called SMT3. The SUMO-conjugating enzyme is called UBC9. The sentrin specific protease (SEPN) cleaves the C-terminal peptide from SUMO which then can bind to E1. | + | [[SUMO]] is a small ubiquitin-like modifier which covalently attaches to cellular proteins to modify their function. SUMO is similar in structure but not in sequence to [[Ubiquitin|ubiquitin]]. In several organisms SUMO is called SMT3. The SUMO-conjugating enzyme is called UBC9. The sentrin specific protease (SEPN) cleaves the C-terminal peptide from SUMO which then can bind to E1. For details on SUMO-1 protein complex see <br /> |
*[[Human SUMO E1 complex]] <br /> | *[[Human SUMO E1 complex]] <br /> | ||
*[[Human SUMO E1 complex with a SUMO1-AMP mimic]]<br /> | *[[Human SUMO E1 complex with a SUMO1-AMP mimic]]<br /> | ||
*[[Human SUMO E1~SUMO1-AMP tetrahedral intermediate mimic]]. | *[[Human SUMO E1~SUMO1-AMP tetrahedral intermediate mimic]]. | ||
| - | |||
| - | {{TOC limit|limit=2}} | ||
| - | |||
| - | |||
| - | {{Clear}} | ||
| - | {{Clear}} | ||
| - | <StructureSection load='3kyc' size='500' frame='true' align='right' scene='3kyc/Cv/1' > | ||
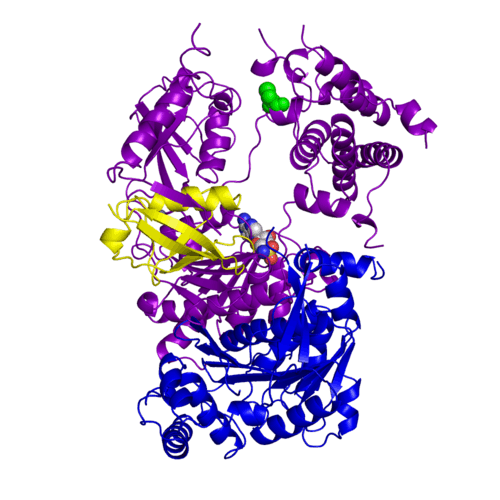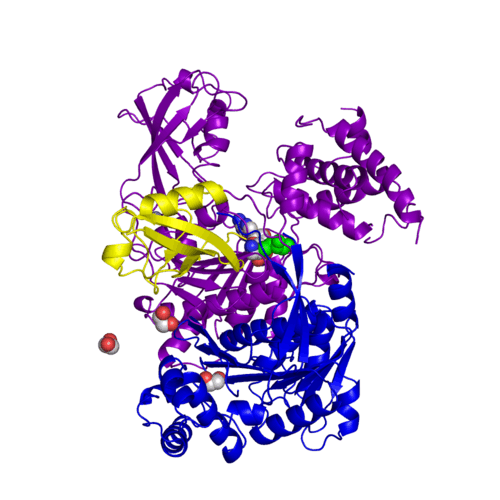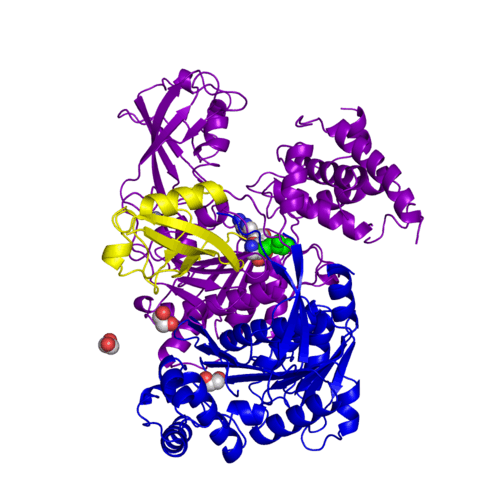
[[Ubiquitin]] (Ub) and ubiquitin-like (Ubl) proteins attached to their target proteins and modulating the activities of those targets in various ways. Three types of evolutionarily conserved enzymes — E1 activating enzymes, E2 conjugating enzymes and E3 ligase enzymes — act sequentially through parallel yet distinct pathways to conjugate ubiquitin and Ubl proteins, such as SUMO and NEDD8, to their targets. The E1 enzyme uses the <scene name='3kyc/Cv/3'>adenosine triphosphate (ATP)</scene> and magnesium to adenylate the C-terminal Ub/Ubl glycine, releasing pyrophosphate and resulting in <scene name='3kyc/Cv/8'>adenosine monophosphate (AMP)</scene>. A non-hydrolysable <scene name='3kyc/Cv/4'>mimic of the acyl adenylate intermediate (AMSN)</scene> and <scene name='3kyc/Cv/5'>mimic of the tetrahedral intermediate (AVSN)</scene> were constructed. In both these compounds the atom of <font color='orange'><b>phosphorus</b></font> is replaced by sulfur (colored <font color='yellow'><b>yellow</b></font>). | [[Ubiquitin]] (Ub) and ubiquitin-like (Ubl) proteins attached to their target proteins and modulating the activities of those targets in various ways. Three types of evolutionarily conserved enzymes — E1 activating enzymes, E2 conjugating enzymes and E3 ligase enzymes — act sequentially through parallel yet distinct pathways to conjugate ubiquitin and Ubl proteins, such as SUMO and NEDD8, to their targets. The E1 enzyme uses the <scene name='3kyc/Cv/3'>adenosine triphosphate (ATP)</scene> and magnesium to adenylate the C-terminal Ub/Ubl glycine, releasing pyrophosphate and resulting in <scene name='3kyc/Cv/8'>adenosine monophosphate (AMP)</scene>. A non-hydrolysable <scene name='3kyc/Cv/4'>mimic of the acyl adenylate intermediate (AMSN)</scene> and <scene name='3kyc/Cv/5'>mimic of the tetrahedral intermediate (AVSN)</scene> were constructed. In both these compounds the atom of <font color='orange'><b>phosphorus</b></font> is replaced by sulfur (colored <font color='yellow'><b>yellow</b></font>). | ||
Revision as of 13:48, 16 July 2013
| |||||||||||
For better understanding of the difference between these two conformations you can see this morph (generated by using POLYVIEW-3D: http://polyview.cchmc.org/polyview3d.html; reload/refresh this page to restart this movie). Of note, in contrast to the previous figure, the same domains of these two structures (3kyc and 3kyd) are colored in the same colors (SUMO1 in yellow, SAE1 colored in blue and other domains in darkviolet). The catalytic Cys173 is shown in the spacefill representation and colored green, AMSN (or AVSN) are shown in the spacefill representation and colored in CPK colors.
Contents |
3D Structures of SUMO
SUMO
2k8h – SUMO – NMR – Trypanosoma brucei
1u4a – hSUMO-3 (mutant) – NMR – human
1a5r - hSUMO-1 - NMR
1wm2, 1wm3, 2awt – hSUMO-2
SUMO+ubiquitin-like SUMO-conjugating enzyme
2vrr, 2uyz – mSUMO-1+Ubc9
2pe6 - hSUMO-1+Ubc9
SUMO+sentrin specific protease
2io0 – pre-hSUMO-2+SEPN2
2io1 - pre-hSUMO-3+SEPN2
2g4d - hSUMO-1+SEPN1
2iy1 - hSUMO-1+SEPN1 (mutant)
2iyd, 2ckh - hSUMO-2+SEPN1
1tgz - hSUMO-1+SEPN2
2io3 - pre-hSUMO-2+SEPN2 (mutant)+RAN GTPase-activating enzyme (mutant)
2iy0 - hSUMO-1+SEPN1 (mutant)+RAN GTPase-activating enzyme
SUMO+ubiquitin-conjugating enzyme
1z5s - hSUMO-1+E2+ RAN GTPase-activating enzyme
2bf8 - SUMO-1+E2 - bovine
SMT3
2k1f – SMT3 – Fruit fly
2eke – ySMT3+UBC9 – yeast
1euv – ySMT3+ULP1 protease
3v60, 3v61 – ySMT3 + PCNA (mutant)
3v62 – ySMT3 + PCNA (mutant) + ATP-dependent DNA helicase SRS2
SUMO+other proteins
2asq – hSUMO-1+SUMO-binding motif in PIASX
3kyc, 3kyd – hSUMO+SUMO-activating enzyme
2rpq – hSUMO-3+activating transcription factor
1wyw - hSUMO-1+thymine DNA glycosylate
2d07 - hSUMO-3+thymine DNA glycosylate
2kqs - hSUMO-1+death domain-associated protein 6 fragment
Reference
- Olsen SK, Capili AD, Lu X, Tan DS, Lima CD. Active site remodelling accompanies thioester bond formation in the SUMO E1. Nature. 2010 Feb 18;463(7283):906-12. PMID:20164921 doi:10.1038/nature08765