Acetylcholinesterase
From Proteopedia
(Difference between revisions)
| Line 6: | Line 6: | ||
Solution of the three-dimensional (3D) structure of [http://en.wikipedia.org/wiki/Pacific_electric_ray ''Torpedo californica''] [[acetylcholinesterase]] (''Tc''AChE) in 1991 opened up new horizons in research on an [http://en.wikipedia.org/wiki/Enzyme enzyme] that had already been the subject of intensive investigation.<ref>PMID:1678899</ref> The unanticipated structure of this extremely rapid enzyme, in which the [http://en.wikipedia.org/wiki/Active_site active site] was found to be buried at the bottom of a <scene name='2ace/Active_site/3'>deep and narrow gorge</scene>, lined by <scene name='2ace/Active_site/4'>14 aromatic residues</scene> <font color='darkmagenta'><b>(colored dark magenta)</b></font>, led to a revision of the views then held concerning [http://en.wikipedia.org/wiki/Substrate_(biochemistry) substrate] traffic, recognition and hydrolysis.<ref>PMID:10545346</ref> To understand how those aromatic residues behave with the enzyme, see [[Flexibility of aromatic residues in acetylcholinesterase]]. Solution of the 3D structure of acetylcholinesterase led to a series of theoretical and experimental studies, which took advantage of recent advances in theoretical techniques for treatment of [http://en.wikipedia.org/wiki/Protein proteins], such as | Solution of the three-dimensional (3D) structure of [http://en.wikipedia.org/wiki/Pacific_electric_ray ''Torpedo californica''] [[acetylcholinesterase]] (''Tc''AChE) in 1991 opened up new horizons in research on an [http://en.wikipedia.org/wiki/Enzyme enzyme] that had already been the subject of intensive investigation.<ref>PMID:1678899</ref> The unanticipated structure of this extremely rapid enzyme, in which the [http://en.wikipedia.org/wiki/Active_site active site] was found to be buried at the bottom of a <scene name='2ace/Active_site/3'>deep and narrow gorge</scene>, lined by <scene name='2ace/Active_site/4'>14 aromatic residues</scene> <font color='darkmagenta'><b>(colored dark magenta)</b></font>, led to a revision of the views then held concerning [http://en.wikipedia.org/wiki/Substrate_(biochemistry) substrate] traffic, recognition and hydrolysis.<ref>PMID:10545346</ref> To understand how those aromatic residues behave with the enzyme, see [[Flexibility of aromatic residues in acetylcholinesterase]]. Solution of the 3D structure of acetylcholinesterase led to a series of theoretical and experimental studies, which took advantage of recent advances in theoretical techniques for treatment of [http://en.wikipedia.org/wiki/Protein proteins], such as | ||
[http://en.wikipedia.org/wiki/Molecular_dynamics molecular dynamics] and [http://en.wikipedia.org/wiki/Electrostatics electrostatics] and to [http://en.wikipedia.org/wiki/Site-directed_mutagenesis site-directed mutagenesis], utilizing suitable expression | [http://en.wikipedia.org/wiki/Molecular_dynamics molecular dynamics] and [http://en.wikipedia.org/wiki/Electrostatics electrostatics] and to [http://en.wikipedia.org/wiki/Site-directed_mutagenesis site-directed mutagenesis], utilizing suitable expression | ||
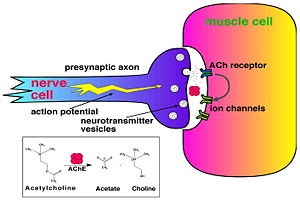
| - | systems. [http://en.wikipedia.org/wiki/Acetylcholinesterase Acetylcholinesterase] [http://en.wikipedia.org/wiki/Hydrolysis hydrolysizes] the [http://en.wikipedia.org/wiki/Neurotransmitter neurotransmitter] [http://en.wikipedia.org/wiki/Acetylcholine acetylcholine] <scene name='2ace/Cv/2'>(ACh)</scene>, producing <scene name='2ace/Cv/3'>choline and an acetate</scene> group. ACh directly binds <scene name='2ace/Cv/4'>Ser200</scene> (via its [http://en.wikipedia.org/wiki/Nucleophile nucleophilic] Oγ atom) within the <scene name='2ace/Cv/5'>catalytic triad (Ser200, His440, and Glu327)</scene> (ACh/''Tc''AChE structure [[2ace]]). The residues <scene name='2ace/Cv/6'>Trp84 and Phe330</scene> are also important in the [http://en.wikipedia.org/wiki/Ligand ligand] recognition <ref name="Raves">PMID:8989325</ref>. After this binding acetylcholinesterase <scene name='2ace/Cv/7'>hydrolysizes</scene> ACh. See also: [[AChE inhibitors and substrates]] | + | systems. [http://en.wikipedia.org/wiki/Acetylcholinesterase Acetylcholinesterase] [http://en.wikipedia.org/wiki/Hydrolysis hydrolysizes] the [http://en.wikipedia.org/wiki/Neurotransmitter neurotransmitter] [http://en.wikipedia.org/wiki/Acetylcholine acetylcholine] <scene name='2ace/Cv/2'>(ACh)</scene>, producing <scene name='2ace/Cv/3'>choline and an acetate</scene> group. ACh directly binds <scene name='2ace/Cv/4'>Ser200</scene> (via its [http://en.wikipedia.org/wiki/Nucleophile nucleophilic] Oγ atom) within the <scene name='2ace/Cv/5'>catalytic triad (Ser200, His440, and Glu327)</scene> (ACh/''Tc''AChE structure [[2ace]]). The residues <scene name='2ace/Cv/6'>Trp84 and Phe330</scene> are also important in the [http://en.wikipedia.org/wiki/Ligand ligand] recognition <ref name="Raves">PMID:8989325</ref>. After this binding acetylcholinesterase <scene name='2ace/Cv/7'>hydrolysizes</scene> ACh. See also:<br /> |
| + | [[AChE inhibitors and substrates]]<br /> | ||
| + | [[Acetylcholinesterase (Hebrew)]] | ||
{{Clear}} | {{Clear}} | ||
Revision as of 10:32, 27 January 2016
| |||||||||||
Contents |
3D Structures of AChE
See 3D structures of acetylcholinesterase
Additional Resources
For additional information, see:
Alzheimer's Disease
AChE inhibitors and substrates
External Links
- Acetylcholinesterase Tutorial by Karl Oberholser, Messiah College
- PDB Molecule of the Month - Acetylcholinesterase
- Movies: X-ray Damage in ACh & Nature's Vacuum Cleaner by R. Gillilan, Cornell Univ
References
- ↑ Sussman JL, Harel M, Frolow F, Oefner C, Goldman A, Toker L, Silman I. Atomic structure of acetylcholinesterase from Torpedo californica: a prototypic acetylcholine-binding protein. Science. 1991 Aug 23;253(5022):872-9. PMID:1678899
- ↑ Botti SA, Felder CE, Lifson S, Sussman JL, Silman I. A modular treatment of molecular traffic through the active site of cholinesterase. Biophys J. 1999 Nov;77(5):2430-50. PMID:10545346
- ↑ 3.0 3.1 Raves ML, Harel M, Pang YP, Silman I, Kozikowski AP, Sussman JL. Structure of acetylcholinesterase complexed with the nootropic alkaloid, (-)-huperzine A. Nat Struct Biol. 1997 Jan;4(1):57-63. PMID:8989325
- ↑ Greenblatt HM, Kryger G, Lewis T, Silman I, Sussman JL. Structure of acetylcholinesterase complexed with (-)-galanthamine at 2.3 A resolution. FEBS Lett. 1999 Dec 17;463(3):321-6. PMID:10606746
- ↑ Harel M, Schalk I, Ehret-Sabatier L, Bouet F, Goeldner M, Hirth C, Axelsen PH, Silman I, Sussman JL. Quaternary ligand binding to aromatic residues in the active-site gorge of acetylcholinesterase. Proc Natl Acad Sci U S A. 1993 Oct 1;90(19):9031-5. PMID:8415649
- ↑ Bar-On P, Millard CB, Harel M, Dvir H, Enz A, Sussman JL, Silman I. Kinetic and structural studies on the interaction of cholinesterases with the anti-Alzheimer drug rivastigmine. Biochemistry. 2002 Mar 19;41(11):3555-64. PMID:11888271
- ↑ Haviv H, Wong DM, Greenblatt HM, Carlier PR, Pang YP, Silman I, Sussman JL. Crystal packing mediates enantioselective ligand recognition at the peripheral site of acetylcholinesterase. J Am Chem Soc. 2005 Aug 10;127(31):11029-36. PMID:16076210 doi:http://dx.doi.org/10.1021/ja051765f
- ↑ 8.0 8.1 Ravelli RB, Raves ML, Ren Z, Bourgeois D, Roth M, Kroon J, Silman I, Sussman JL. Static Laue diffraction studies on acetylcholinesterase. Acta Crystallogr D Biol Crystallogr. 1998 Nov 1;54(Pt 6 Pt 2):1359-66. PMID:10089512
- ↑ 9.0 9.1 Harel M, Sonoda LK, Silman I, Sussman JL, Rosenberry TL. Crystal structure of thioflavin T bound to the peripheral site of Torpedo californica acetylcholinesterase reveals how thioflavin T acts as a sensitive fluorescent reporter of ligand binding to the acylation site. J Am Chem Soc. 2008 Jun 25;130(25):7856-61. Epub 2008 May 31. PMID:18512913 doi:http://dx.doi.org/10.1021/ja7109822
- ↑ Greenblatt HM, Guillou C, Guenard D, Argaman A, Botti S, Badet B, Thal C, Silman I, Sussman JL. The complex of a bivalent derivative of galanthamine with torpedo acetylcholinesterase displays drastic deformation of the active-site gorge: implications for structure-based drug design. J Am Chem Soc. 2004 Dec 1;126(47):15405-11. PMID:15563167 doi:http://dx.doi.org/10.1021/ja0466154
- ↑ Kryger G, Silman I, Sussman JL. Structure of acetylcholinesterase complexed with E2020 (Aricept): implications for the design of new anti-Alzheimer drugs. Structure. 1999 Mar 15;7(3):297-307. PMID:10368299
- ↑ Sanson B, Nachon F, Colletier JP, Froment MT, Toker L, Greenblatt HM, Sussman JL, Ashani Y, Masson P, Silman I, Weik M. Crystallographic Snapshots of Nonaged and Aged Conjugates of Soman with Acetylcholinesterase, and of a Ternary Complex of the Aged Conjugate with Pralidoxime (dagger). J Med Chem. 2009 Jul 30. PMID:19642642 doi:10.1021/jm900433t
- ↑ Paz A, Roth E, Ashani Y, Xu Y, Shnyrov VL, Sussman JL, Silman I, Weiner L. Structural and functional characterization of the interaction of the photosensitizing probe methylene blue with Torpedo californica acetylcholinesterase. Protein Sci. 2012 Jun 1. doi: 10.1002/pro.2101. PMID:22674800 doi:10.1002/pro.2101
Treatments:AChE Inhibitor References
Treatments:Alzheimer's Disease
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Joel L. Sussman, Alexander Berchansky, David Canner, Eran Hodis, Clifford Felder, Jaime Prilusky, Harry Greenblatt, Yechun Xu