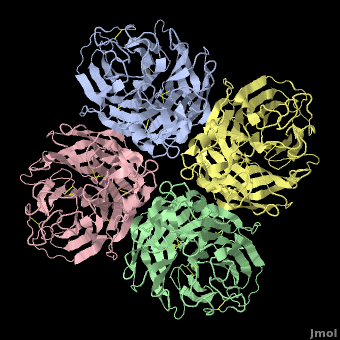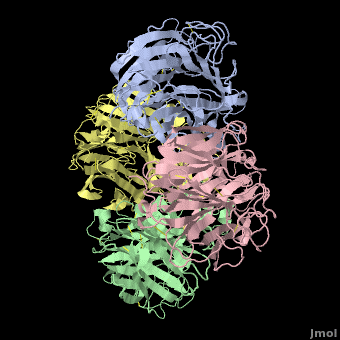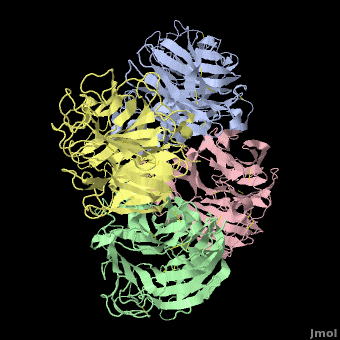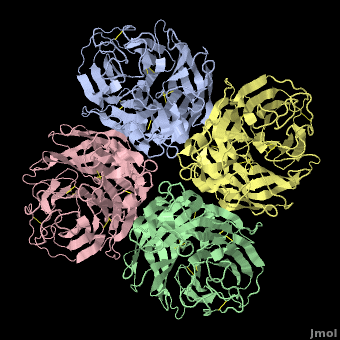Avian Influenza Neuraminidase, Tamiflu and Relenza
From Proteopedia
(Difference between revisions)
| Line 88: | Line 88: | ||
====Tamiflu Binds to N1 by Induced Fit==== | ====Tamiflu Binds to N1 by Induced Fit==== | ||
| - | <scene name='Avian_Influenza_Neuraminidase,_Tamiflu_and_Relenza/Morph_2hty_to_2hu4/2'>Morph of N1 alone to N1 complexed with Tamiflu. </scene> The position where Tamiflu will bind is shown translucent except when bound in the empirically-determined model. N1 alone - ([[2hty]]), N1 complexed with Tamiflu - ([[2hu4]]). | + | <scene name='Avian_Influenza_Neuraminidase,_Tamiflu_and_Relenza/Morph_2hty_to_2hu4/2'>Morph of N1 alone to N1 complexed with Tamiflu. </scene> The position where Tamiflu will bind is shown translucent except when bound in the empirically-determined model. N1 alone - ([[2hty]]), N1 complexed with Tamiflu - ([[2hu4]]). If the morph does not animate automatically, click this button: {{Template:Button Toggle Animation2}} |
Tamiflu was designed to fit N2/N9, so it is serendipitous that it works on N1. In fact, when the structure of N1 was determined<ref name='Russell2006' />, the <font color='#e07000'><b>loop comprising residues 147-152</b></font> was not in a suitable position to participate in binding Tamiflu. However, the complex of N1 with Tamiflu revealed that this loop is pulled into proper contact with the drug in an [[Induced fit|induced fit]] manner<ref name='Russell2006' />. A [[Morphs|morph]] from N1 alone ([[2hty]]) to N1 complexed with Tamiflu ([[2hu4]])<ref>Chain A from [[2hty]] was [[morphs|morphed]] to chain A of [[2hu4]] by linear interpolation, inserting 6 intermediate interpolated frames, using the freely available [http://www.umass.edu/microbio/rasmol/pdbtools.htm#martz morph2 program].</ref> shows the change in position of this loop (<scene name='Avian_Influenza_Neuraminidase,_Tamiflu_and_Relenza/Morph_2hty_to_2hu4/8'>replay initial morph</scene>). | Tamiflu was designed to fit N2/N9, so it is serendipitous that it works on N1. In fact, when the structure of N1 was determined<ref name='Russell2006' />, the <font color='#e07000'><b>loop comprising residues 147-152</b></font> was not in a suitable position to participate in binding Tamiflu. However, the complex of N1 with Tamiflu revealed that this loop is pulled into proper contact with the drug in an [[Induced fit|induced fit]] manner<ref name='Russell2006' />. A [[Morphs|morph]] from N1 alone ([[2hty]]) to N1 complexed with Tamiflu ([[2hu4]])<ref>Chain A from [[2hty]] was [[morphs|morphed]] to chain A of [[2hu4]] by linear interpolation, inserting 6 intermediate interpolated frames, using the freely available [http://www.umass.edu/microbio/rasmol/pdbtools.htm#martz morph2 program].</ref> shows the change in position of this loop (<scene name='Avian_Influenza_Neuraminidase,_Tamiflu_and_Relenza/Morph_2hty_to_2hu4/8'>replay initial morph</scene>). | ||
| - | The binding of Tamiflu to N1 pulls the sidechains of two conserved residues, <scene name='Avian_Influenza_Neuraminidase,_Tamiflu_and_Relenza/Morph_2hty_to_2hu4/7'>Asp151, Glu119</scene> (<font color="red">Sorry, this scene is temporarily broken.</font>), closer to the inhibitor. | + | The binding of Tamiflu to N1 pulls the sidechains of two conserved residues, <scene name='Avian_Influenza_Neuraminidase,_Tamiflu_and_Relenza/Morph_2hty_to_2hu4/7'>Asp151, Glu119</scene><!--(<font color="red">Sorry, this scene is temporarily broken.</font>)-->, closer to the inhibitor. |
===Cavity in N1: An Opportunity for Drug Design=== | ===Cavity in N1: An Opportunity for Drug Design=== | ||
Current revision
| |||||||||||
Links
- cdc.gov/flu, the official influenza resource of the US Center for Disease Control.
- Pandemic Planning Toolkit (by Roche).
- Relenza offical website by GlaxoSmithKline.
- Tamiflu official website by Roche.
- The Next Pandemic? An authoritative overview of economic and political factors written in 2005 by Laurie Garrett.
Notes and Literature References
- ↑ See Influenza A Types of Influenza Virus (in Wikipedia).
- ↑ Epidemiology of Avian Influenza at the Food and Agriculture Organization of the United Nations.
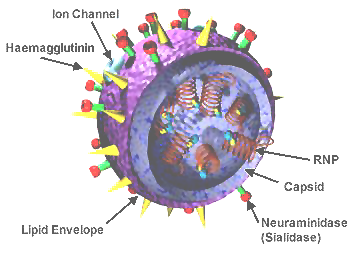
- ↑ Image of influenza virus structure was obtained from Wikipedia.
- ↑ Influenza at Wikipedia.
- ↑ The tetramer is one of two biological units in the asymmetric unit of 2hu4.
- ↑ See Evolutionary Conservation. Coloring by ConSurf on chain A of 2hu4 based on 100 unique homologs using default conditions, done on September 23, 2008.
- ↑ Severe acute respiratory syndrome, SARS, was a near-pandemic in 2002-2003: see Severe Acute Respiratory Syndrome (in Wikipedia).
- ↑ 2009 Swine Flu Outbreak (in Wikipedia).
- ↑ Human Swine Influenza Investigation, April 23, 2009.
- ↑ From the CDC: Investigation and Interim Recommendations: Swine Influenza (H1N1).
- ↑ CDC document on Antiviral Drugs and Swine Influenza.
- ↑ Monto AS. Epidemiology of influenza. Vaccine. 2008 Sep 12;26 Suppl 4:D45-8. PMID:19230159
- ↑ 13.0 13.1 Some immunity to novel H1N1 flu found in seniors (May 21, 2009, University of Minnesota Center for Infectious Disease and Policy).
- ↑ 14.0 14.1 Morbidity and Mortality Weekly Report for May 22, 2009 from the US Centers for Disease Control and Prevention.
- ↑ Influenza Pandemic at Wikipedia.
- ↑ Vaccination for Influenza at Wikipedia
- ↑ What You Need to Know About 2012-2013 Flu Vaccine.
- ↑ Report on amantadine resistance, CDC Morbidity and Mortality Weekly Reports, January 2006.
- ↑ 19.0 19.1 19.2 19.3 Russell RJ, Haire LF, Stevens DJ, Collins PJ, Lin YP, Blackburn GM, Hay AJ, Gamblin SJ, Skehel JJ. The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design. Nature. 2006 Sep 7;443(7107):45-9. Epub 2006 Aug 16. PMID:16915235 doi:http://dx.doi.org/10.1038/nature05114
- ↑ Varghese JN, Laver WG, Colman PM. Structure of the influenza virus glycoprotein antigen neuraminidase at 2.9 A resolution. Nature. 1983 May 5-11;303(5912):35-40. PMID:6843658
- ↑ 21.0 21.1 Collins PJ, Haire LF, Lin YP, Liu J, Russell RJ, Walker PA, Skehel JJ, Martin SR, Hay AJ, Gamblin SJ. Crystal structures of oseltamivir-resistant influenza virus neuraminidase mutants. Nature. 2008 Jun 26;453(7199):1258-61. Epub 2008 May 14. PMID:18480754 doi:http://dx.doi.org/10.1038/nature06956
- ↑ Holmes EC. Virology. Helping the resistance. Science. 2010 Jun 4;328(5983):1243-4. PMID:20522766 doi:10.1126/science.1190994
- ↑ Bloom JD, Gong LI, Baltimore D. Permissive secondary mutations enable the evolution of influenza oseltamivir resistance. Science. 2010 Jun 4;328(5983):1272-5. PMID:20522774 doi:10.1126/science.1187816
- ↑ Layne SP, Monto AS, Taubenberger JK. Pandemic influenza: an inconvenient mutation. Science. 2009 Mar 20;323(5921):1560-1. PMID:19299601 doi:http://dx.doi.org/323/5921/1560
- ↑ Chain A from 2hty was morphed to chain A of 2hu4 by linear interpolation, inserting 6 intermediate interpolated frames, using the freely available morph2 program.
Proteopedia Page Contributors and Editors (what is this?)
Eric Martz, Eran Hodis, David Canner, Ilan Samish, Alexander Berchansky, Jaime Prilusky, David S. Goodsell, Michal Harel, Michael Strong