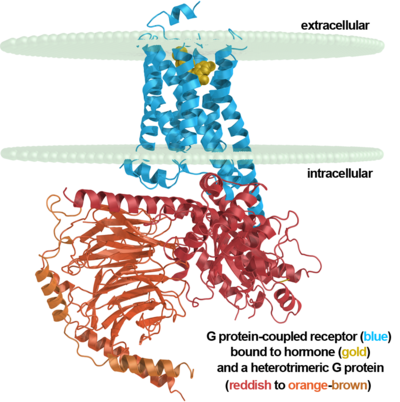
G protein-coupled receptor
From Proteopedia
| (15 intermediate revisions not shown.) | |||
| Line 4: | Line 4: | ||
Rhodopsin shares similar membrane topology with the members of the superfamily, specifically family A of the [[G protein-coupled receptor|G protein-coupled receptors]] which include the seven transmembrane helices, an extracellular N-terminus and cytoplasmic C-terminus<ref name="rhodopsin">PMID:15251227</ref>. | Rhodopsin shares similar membrane topology with the members of the superfamily, specifically family A of the [[G protein-coupled receptor|G protein-coupled receptors]] which include the seven transmembrane helices, an extracellular N-terminus and cytoplasmic C-terminus<ref name="rhodopsin">PMID:15251227</ref>. | ||
| + | |||
| + | See also | ||
| + | *[[CAMP-dependent pathway]] | ||
| + | *[[Cytokine receptors]] | ||
| + | *[[Neuropeptides]] | ||
| + | *[[Transmembrane (cell surface) receptors]] | ||
[[Image:7tm labeled.png|right|400px]] | [[Image:7tm labeled.png|right|400px]] | ||
| Line 39: | Line 45: | ||
* [[Belsomra]] and Orexin receptors | * [[Belsomra]] and Orexin receptors | ||
* [[Hypocretin and receptors]] | * [[Hypocretin and receptors]] | ||
| + | ====[[Oxytocin receptor]]==== | ||
===Subfamily A10=== | ===Subfamily A10=== | ||
| Line 46: | Line 53: | ||
====Human GPR40 (hGPR40), also known as Free Fatty Acid Receptor 1 (FFAR1)==== | ====Human GPR40 (hGPR40), also known as Free Fatty Acid Receptor 1 (FFAR1)==== | ||
* [[GPR40]] | * [[GPR40]] | ||
| - | Like most G-protein coupled receptors, hGPR40 contains <scene name='72/721541/Top_view_transmembrane_helices/2'>7 transmembrane helices</scene> (<scene name='72/721541/Top_view_transmembrane_helices/1'>top view of TM helices</scene>). To obtain a crystal structure of the protein, 4 <scene name='72/721541/Stabilizing_mutations/4'>stabilizing mutations</scene> (<scene name='72/721541/L42a/3'>L42A</scene>, <scene name='72/721541/F88a/4'>F88A</scene>, <scene name='72/721541/G103a/3'>G103A</scene>, <scene name='72/721541/Y202f/3'>Y202F</scene>) were made to increase the expression and thermal stability of the protein. These mutations did not significantly impact the enzyme's binding affinity with a known agonist TAK-875. <scene name='72/721541/Lysozyme_crimson/2'>T4 Lysozyme</scene> (in crimson) was also added to intracellular loop 3 to aid in the formation of crystals. | ||
| - | |||
| - | While there is relatively low sequence identity between hGPR40 and peptide-binding and opioid GPCRs, they do share structural similarities such as a conserved <scene name='72/727085/Hairpin_loop/4'>hairpin loop</scene> motif on <scene name='72/727085/Ecl2/4'>extracellular loop 2 </scene>(ECL2). In addition, a conserved <scene name='72/727085/Disulfide/3'>disulphide bond</scene> is formed between transmembrane helix 3 (Cys 79) and the C-terminus of ECL2 (Cys170). Compared to peptide-binding and opioid GPCRs, which have distinctive β-sheets spanning from transmembrane helix 4 to 5, hGPR40 possesses a shorter B-sheet-like region, which has low B-factors. This reflects the low mobility of the region that limits the overall flexibility of the adjacent portion of ECL2 between Leu171 and Asp175. A unique feature of hGPR40 is the presence of an additional 13 residues (Pro147 to Gly159) on ECL2, which is absent on all the other peptide/opioid receptors. These extra residues form a separate <scene name='72/727085/Auxiliary_loop/3'>auxiliary loop</scene> between the B-sheet-like region and transmembrane 4. Together, the auxiliary loop and ECL2 of hGPR40 function as a <scene name='72/727085/Ecl2_cap/3'>roof </scene> over the canonical binding site covering it from the central extracellular region. | ||
| - | |||
| - | The canonical binding pocket for many other GPCRs is solvent exposed and centrally located between the transmembrane helices allowing ligands to directly bind from the extracellular space. However, because <scene name='72/727085/Ecl2/4'>ECL2</scene> acts as a roof to this canonical binding site, it inhibits ligands from entering directly from the extracellular region. Instead, the highly lipophilic nature of hGPRC40’s ligands allow it to enter a <scene name='72/727085/Hgpr40_entry/2'>noncanonical binding pocket </scene> between TM3 and TM4 by moving through the lipid bilayer. | ||
| - | |||
| - | FFAs bind to hGPR40 by coordinating its free carboxyl group to 3 amino acids, <scene name='72/727085/Ffa_binding/1'>Arg183, Tyr2240, and Arg258</scene>, which are located close to the <scene name='72/727085/Hgpr40_transmane_active/1'>extracellular domain</scene> of hGPR40 on TM5, 6 and 7. Because of the close proximity of these residues to the extracellular domain and the dominantly hydrophobic nature of FFA’s, it is likely that ligand binding occurs close to the plane of the membrane. | ||
| - | |||
| - | The <scene name='72/721541/Tak_binding_site/4'>binding site for the partial agonist TAK-875</scene> has been identified, but other binding sites were hypothesized. TAK-875 binds between transmembrane helices 3, 4, and 5 and underneath ECL2. hGPR40 has a distinct binding pocket that is established by <scene name='72/721541/All_binding_residues/3'>8 key residues</scene>: <scene name='72/721541/Tyr91/1'>Tyr91</scene>, <scene name='72/721541/Glu172/2'>Glu172</scene>, <scene name='72/721541/Arg183/2'>Arg183</scene>, <scene name='72/721541/Ser187/2'>Ser187</scene>, <scene name='72/721541/Tyr240/1'>Tyr240</scene>, <scene name='72/721541/Asn241/1'>Asn241</scene>, <scene name='72/721541/Asn244/1'>Asn244</scene>, and <scene name='72/721541/Arg258/1'>Arg258</scene> (all individual residues shown in chartreuse). The importance of these residues for agonist binding was determined by alanine site-directed-mutagenesis mutagenesis studies. When the substrate (an agonist) enters the binding pocket, 4 of the 8 <scene name='72/721541/Hydrogen_binding_1/8'>key binding residues</scene> interact directly with the carboxylate moiety of the agonist by hydrogen bonding to it. These residues include 2 key arginines in the binding pocket, Arg183 and Arg258, and 2 key tyrosine residues, Tyr91 and Tyr240. Tyr240 is especially important for binding, as mutation of Tyr240 caused an eight fold reduction in the binding affinity of TAK-875 and had a significant effect on the binding affinity (K<sub>D</sub>) of the protein. | ||
| - | |||
| - | hGPR40 contains a highly conserved hairpin extracellular loop (<scene name='72/721541/Ecl2/4'>ECL2</scene>) is the longest and most divergent of the extracellular loops found in proteins (<scene name='72/721541/Ecl2_top/2'>top view of ECL2</scene>). The loop is accompanied by a disulfide bond (<scene name='72/721541/Cysteine_bridge/3'>Cys79 and Cys170</scene>) that forms between transmembrane helix 4 and the C-terminus of the ECL2 loop. In hGPR40, ECL2 has two sections: a beta sheet and an auxiliary loop. The β-sheet spans helices 4 and 5 and is shorter in hGPR40 than in other GPCRs. The ECL2 of hGPR40 also differs from that of other proteins because it contains an auxiliary loop of 13 extra residues. The entire extracellular loop has low mobility and flexibility, which allows it to act as a cap for the binding pocket. The only exception to the low flexibility is the tip of the auxiliary loop, which corresponds to residues Asp152-Asn155. This area of greater mobility allows for substrates to enter the binding site. | ||
| - | |||
| - | <scene name='72/727085/Hgpr40_begin/3'>Tak-875</scene> is a partial agonist of GPR40 and tested for the treatment of type 2 diabetes. The binding of TAK-875 to hGPR40 occurs by the ligand entering the binding site through the membrane bilayer. This membrane insertion is performed via a method similar to ligand binding to sphingosine 1-phosphate receptor 1, retinal loading of GPCR opsin, and the entry of anandamide in cannabinoid receptors, in which the <scene name='72/727085/Ecl2/4'>extracellular loops</scene> block the binding from the extracellular matrix <ref>PMID:22344443</ref>. | ||
| - | |||
| - | TAK-875 binds to the <scene name='72/727085/Hgpr40_entry/2'>noncanonical binding site </scene> created between transmembrane (TM) domains 3-5 and the extracellular loop 2 (ECL2) of hGPR40. The ECL2 and auxiliary loop form a roof causing TAK-875 to enter through TM3 and TM4, first passing through the lipid bilayer. The carboxylate of TAK-875 is buried within a very hydrophobic region and in a complex complex <scene name='72/727085/Hgpr40_binding_relay/6'>charge network</scene> involving Glu172, Ser187, Asn241, and Asn 244 from hGPR40 forming ionic and polar interactions by coordinating TAK-875 with Arg183, Arg258, Tyr91, and Tyr240. | ||
| - | ===Subfamily A13=== | ||
| - | ====Lysophosphatidic acid receptor==== | ||
| - | * [[Lysophosphatidic acid receptor]] | ||
| - | |||
| - | LPA<sub>1</sub> lies in the membrane as shown by the <scene name='72/721545/Membrane/6'>fatty acid</scene> bound in the crystallization of LPA<sub>1</sub> in orange. Most <scene name='72/721545/Polarity/4'>polar amino acids</scene> (red) reside on the intracellular and extracellular areas of the receptor, while most residues positioned on the trans membrane helices inside the membrane are hydrophobic (blue). A cytochrome b (b<sub>562</sub>RIL) protein was inserted into the 3rd intracellular loop to facilitate crystallization. The intracellular region of this membrane protein is coupled to a heterotrimeric G protein. | ||
| - | |||
| - | Three native <scene name='72/721545/Disulfides/5'>disulfide bonds</scene> in the extracellular region of this receptor provide fold stability. The 1st disulfide bond constrains the N terminal helix to extracellular loop (ECL) 2. The 2nd disulfide bond shapes ECL2, and the 3rd binds ECL3 to one of the transmembrane alpha helices. These disulfide bonds provide intramolecular stabilization along the extracellular region of the LPA<sub>1</sub> receptor, where the substrate enters into the binding pocket. The <scene name='72/721545/N-terminus/3'>N-terminus</scene> is a 6 turn α-helix and functions like a cap on the extracellular side of the protein, packing tightly against ECL1 and ECL2. The N-terminus helix also provides <scene name='72/721545/34_39_40/4'>polar amino acids</scene> that interact with the ligand when bound. The extracellular region of this receptor plays a role in substrate specificity. | ||
| - | |||
| - | The biological ligand of the LPA<sub>1</sub> receptor receptor is lysophosphatidic acid (LPA), a phospholipid that contains a long, nonpolar tail, a phosphate head, a chiral hydroxyl group, and an ester group. This receptor provides specificity for its ligand by the amphipathic binding pocket; the positive region on the left hand side of the pocket stabilizes the LPA's phosphate group, the nonpolar region at the bottom of the binding pocket stabilizes the hydrophobic tail of LPA, and the polar region at the top of the pocket stabilize binding of the ester and hydroxyl group. The <scene name='72/721545/Ligand/4'>binding pocket</scene> for LPA consists of both polar and nonpolar residues. <scene name='72/721545/All_polar_interactions/7'>Polar</scene> residues are located on the N terminus and within the binding pocket. A <scene name='72/721545/Hydrophobic_pocket/4'>hydrophobic pocket</scene> also interacts with the long acyl chain of LPA. The shape and polarity of the binding pocket makes it specific for molecules with a polar head and long hydrophobic tail shaped like LPA. ONO-9780307 (ON7) is an antagonist for LPA due to its large nonpolar region, chiral hydroxyl group, ester, and carboxylic acid which all resemble portions of the LPA molecule. 4 separate interactions with this antagonist of LPA<sub>1</sub> help demonstrate the key interactions that stabilize the binding of the LPA phospholipid to this receptor. In the nonpolar region of the binding pocket, <scene name='72/721543/Nonpolar/2'>three non polar residues</scene> of LPA<sub>1</sub> stabilize the large nonpolar group of ON7. At the polar region, the ligand binding is stabilized by <scene name='72/721543/Arg124gln125/4'>Arg124 and Glu125</scene> forming ionic and polar interactions with the carboxylic acid and the hydroxyl group of ON7. In addition, interplay between <scene name='72/721543/Lys39_and_glu293/8'>Glu293 and Lys39</scene> causes another stabilizing component with the ON7 antagonist. Glu293 forms polar interactions with Lys39, positioning it in close proximity to to the carboxylic acid of ON7, which then interactions with Lys39 via ionic bonding. While Lys39 is highly conserved among all 6 LPA receptors, a neighboring His residue is specific to the LPA<sub>1</sub> receptor. <scene name='72/721543/His40/4'>His40</scene> forms both ionic and polar interactions with the carboxylic acid of ON7. Protonation of this residue greatly affects the binding affinity of LPA, leading to an increase in the pathways associated with cell proliferation and migration. Because cancerous tumors create acidic environments where His40 is protonated, this residue is an important link to tumor growth and cancer cell movement. | ||
====Sphingosine 1-phosphate Receptor==== | ====Sphingosine 1-phosphate Receptor==== | ||
| Line 102: | Line 85: | ||
**Noradrenaline | **Noradrenaline | ||
**Carmoterol, see [[2y02]]. | **Carmoterol, see [[2y02]]. | ||
| - | **Salbutamol (Albuterol in USA), [[2y04]]. | + | **[[Salbutamol]] (Albuterol in USA), [[2y04]]. |
*Beta blockers: | *Beta blockers: | ||
**Metoprolol | **Metoprolol | ||
**Atenolol | **Atenolol | ||
**Bisoprolol | **Bisoprolol | ||
| - | **Propranolol | + | **[[Propranolol]] |
| - | **Timolol | + | **[[Timolol]] |
**Nebivolol | **Nebivolol | ||
**Vortioxetine | **Vortioxetine | ||
| Line 121: | Line 104: | ||
*G<sub>s</sub>: adenylate cyclase activated, cAMP up. For G<sub>s</sub> see [[Beta2 adrenergic receptor-Gs protein complex updated]]. | *G<sub>s</sub>: adenylate cyclase activated, cAMP up. For G<sub>s</sub> see [[Beta2 adrenergic receptor-Gs protein complex updated]]. | ||
β2-adrenergic agonists: | β2-adrenergic agonists: | ||
| - | **Salbutamol (Albuterol in USA) | + | **[[Salbutamol]] (Albuterol in USA) |
**Bitolterol mesylate | **Bitolterol mesylate | ||
| - | **Formoterol | + | **[[Formoterol]] |
**Isoprenaline | **Isoprenaline | ||
**Levalbuterol | **Levalbuterol | ||
**Metaproterenol | **Metaproterenol | ||
| - | **Salmeterol | + | **[[Salmeterol]] |
**Terbutaline | **Terbutaline | ||
**Ritodrine | **Ritodrine | ||
*Beta blockers: | *Beta blockers: | ||
**Butoxamine | **Butoxamine | ||
| - | **Timolol | + | **[[Timolol]] |
| - | **Propranolol | + | **[[Propranolol]] |
**ICI-118,551 | **ICI-118,551 | ||
**Paroxetine | **Paroxetine | ||
| Line 188: | Line 171: | ||
==Family C of GPCRs, Metabotropic glutamate receptors== | ==Family C of GPCRs, Metabotropic glutamate receptors== | ||
| + | Metabotropic glutamate receptors are [[Glutamate Receptors|glutamate receptors]] that activate ion channels indirectly through a signaling cascade involving G proteins<ref>PMID:20716669</ref>. Glutamate receptors are classified into 3 groups based on their homology, mechanism and pharmacological properties. | ||
*[[Metabotropic glutamate receptor|Metabotropic Glutamate Receptors]] | *[[Metabotropic glutamate receptor|Metabotropic Glutamate Receptors]] | ||
*[[Ligand Binding N-Terminal of Metabotropic Glutamate Receptors]] | *[[Ligand Binding N-Terminal of Metabotropic Glutamate Receptors]] | ||
*[[Metabotropic glutamate receptor 5]] | *[[Metabotropic glutamate receptor 5]] | ||
| + | |||
| + | ==Metabotropic GABA receptors (GABAB)== | ||
| + | GABAB receptors (GABABR) are G-protein coupled receptors for gamma-aminobutyric acid (GABA), therefore making them metabotropic receptors, that are linked via G-proteins to potassium channels. GABAB receptors also reduces the activity of adenylyl cyclase and Ca2+ channels by using G-proteins with Gi/G0 α subunits. | ||
| + | *[[GABA receptor]] | ||
| + | *[[User:Rana Saad/The human GABAb receptor]] | ||
=Nobel Prize Related to the Structures= | =Nobel Prize Related to the Structures= | ||
Current revision
G protein-coupled receptors, often abbreviated GPCRs, are an abundant superfamily of proteins also known as seven-transmembrane domain receptors, 7TM receptors, 7 pass transmembrane receptors, heptahelical receptors, serpentine receptor, and G protein-linked receptors (GPLRs). G protein-coupled receptors are cell surface signalling proteins involved in many physiological functions and in multiple diseases. They are also the target of the majority of all modern medicinal drugs[1][2]. The extracellular side is generally where the ligand enters for binding. On the intracellular side they interact with G proteins involved in signaling induced by the binding of the ligand.
Illustrating their importance and the largesse of the superfamily, there are roughly 800 known members of the superfamily in the human genome alone — estimated to be 4% of human protein-coding genes. Members are further subclassified into one of five families of GPCRs[3].
Rhodopsin shares similar membrane topology with the members of the superfamily, specifically family A of the G protein-coupled receptors which include the seven transmembrane helices, an extracellular N-terminus and cytoplasmic C-terminus[4].
See also
Contents
|
List of the G protein-coupled receptors
Family A of GPCRs
Neurotensin receptor
Subfamily A2
CXC Chemokine receptors
Subfamily A4
Opioid receptors
Opioid receptor (OpR) is a G protein-coupled receptor with opioids as ligands[5]. OpR types are classified according to the ligands which bind to them.
- The μ-opioid receptor binds morphine. For more details on μ-opioid receptor see
- Mu Opioid Receptor Bound to a Morphinan Antagonist
- μ Opioid Receptors
- Mu Opioid Receptor.
- 6dde μ-opioid receptor: G protein complex
- The κ-opioid receptor binds opium-type ligands. For details see Student Project 3 for UMass Chemistry 423 Spring 2015.
- The δ-opioid receptor binds enkephalins. For details see Delta opioid receptor.
- The Nociceptin/orphanin FQ opioid receptor binds the heptadecapeptide orphanin[6].
See also
Subfamily A6
Orexin receptor
- Orexin and Orexin receptor
- Belsomra and Orexin receptors
- Hypocretin and receptors
Oxytocin receptor
Subfamily A10
Subfamily A11
Human GPR40 (hGPR40), also known as Free Fatty Acid Receptor 1 (FFAR1)
Sphingosine 1-phosphate Receptor
The sphingosine-1-phosphate receptors are a class of G protein-coupled receptors that are targets of the lipid signalling molecule Sphingosine-1-phosphate (S1P).
See User:Harish Srinivas/Sandbox 1
Subfamily A16
Rhodopsins
The product of light activation, Metarhodopsin II, initiates the visual phototransduction pathway by stimulating the G protein Transducin (Gt), resulting in the liberation of its α subunit. This GTP-bound subunit in turn activates cGMP phosphodiesterase. cGMP phosphodiesterase hydrolyzes (breaks down) cGMP, lowering its local concentration so it can no longer activate cGMP-dependent cation channels. Phosphodiesterase 6 is the primary effector of retinal phototransduction. See details in User:Rick H. Cote/PDE6.
Subfamily A17
5-Hydroxytryptamine (5-HT) receptors (Serotonin receptors)
Adrenergic receptors
β1 adrenergic receptor
- 3D structures in Adrenergic receptor.
- Beta-1 Adrenergic receptor
- Gs: adenylate cyclase activated, cAMP up.
- β1-adrenergic agonists:
- Dobutamine, see Beta-1 Adrenergic receptor, 2y00, 2y01, 6h7l.
- Isoprenaline, see Beta-1 Adrenergic receptor, 2y03.
- Noradrenaline
- Carmoterol, see 2y02.
- Salbutamol (Albuterol in USA), 2y04.
- Beta blockers:
- Metoprolol
- Atenolol
- Bisoprolol
- Propranolol
- Timolol
- Nebivolol
- Vortioxetine
β2 adrenergic receptor
| |||||||||||
| |||||||||||
- The human β2 adrenergic receptor bound to a G-protein (3sn6) is featured in a scene above, and additional structures are on the Adrenergic receptor page.
- Article Beta-2 Adrenergic Receptor by Wayne Decatur, David Canner, Dotan Shaniv, Joel L. Sussman, Michal Harel
- Article Beta-2 adrenergic receptor by Joel L. Sussman, Tala Curry, Michal Harel, Jaime Prilusky
- Group:SMART:A Physical Model of the beta-Adrenergic Receptor
- Gs: adenylate cyclase activated, cAMP up. For Gs see Beta2 adrenergic receptor-Gs protein complex updated.
β2-adrenergic agonists:
- Salbutamol (Albuterol in USA)
- Bitolterol mesylate
- Formoterol
- Isoprenaline
- Levalbuterol
- Metaproterenol
- Salmeterol
- Terbutaline
- Ritodrine
- Beta blockers:
- Butoxamine
- Timolol
- Propranolol
- ICI-118,551
- Paroxetine
Dopamine Receptor
There are five subtype dopamine receptors, D1, D2, D3, D4, and D5. The D3 receptor is a part of the D2-like family.[7] The D2-like family receptors are coupled to the G protein Giα, which directly inhibits the formation of cAMP by inhibiting the enzyme adenylyl cyclase.
Subfamily A18
Histamine receptors
The H1 receptor is a histamine receptor belonging to the family of rhodopsin-like G-protein-coupled receptors. The H1 receptor is linked to an intracellular G-protein (Gq) that activates phospholipase C (see Unique bidirectional interactions of Phospholipase C beta 3 with G alpha Q and the inositol triphosphate (IP3) signalling pathway. When a ligand binds to a G protein-coupled receptorthat is coupled to a Gq heterotrimeric G protein, the α-subunit of Gq can bind to and induce activity in the PLC isozyme PLC-β, which results in the cleavage of PIP2 into IP3 and DAG.
- Histamine H1 receptor
- 3rze - human histamine H1 receptor with an antagonist doxepin.
Adenosine A2A receptor
Gs → cAMP up
- Adenosine A2A receptor
- Effect of Caffeine (Trimethylxanthine) on Human A2A Receptor
- Adenosine A2A receptor 3D structures
Agonists:
- N6-3-methoxyl-4-hydroxybenzyl adenine riboside (B2)
- ATL-146e
- CGS-21680
- Regadenoson
- Adenosine
Antagonists:
- Caffeine
- aminophylline
- theophylline
- istradefylline
- SCH-58261
- SCH-442,416
- ZM-241,385
Muscarinic acetylcholine receptors
M1, M3, M5 receptors are coupled with Gq proteins, while M2 and M4 receptors are coupled with Gi/o proteins.
Family B of GPCRs
These receptors activate adenylyl cyclase and the phosphatidyl-inositol-calcium pathway. The glucagon receptor is a 62 kDa protein that is activated by glucagon and is a member of the class B G-protein coupled family of receptors, coupled to G alpha i, Gs and to a lesser extent G alpha q. Stimulation of the receptor results in the activation of adenylate cyclase and phospholipase C and in increased levels of the secondary messengers intracellular cAMP and calcium.
Subfamily B1
Glucose-dependent Insulinotropic Polypeptide Receptor
Glucagon receptor
Glucagon-like peptide 1 receptor
Family C of GPCRs, Metabotropic glutamate receptors
Metabotropic glutamate receptors are glutamate receptors that activate ion channels indirectly through a signaling cascade involving G proteins[8]. Glutamate receptors are classified into 3 groups based on their homology, mechanism and pharmacological properties.
- Metabotropic Glutamate Receptors
- Ligand Binding N-Terminal of Metabotropic Glutamate Receptors
- Metabotropic glutamate receptor 5
Metabotropic GABA receptors (GABAB)
GABAB receptors (GABABR) are G-protein coupled receptors for gamma-aminobutyric acid (GABA), therefore making them metabotropic receptors, that are linked via G-proteins to potassium channels. GABAB receptors also reduces the activity of adenylyl cyclase and Ca2+ channels by using G-proteins with Gi/G0 α subunits.
Nobel Prize Related to the Structures
Robert J. Lefkowitz and Brian K. Kobilka share the 2012 Nobel Prize in Chemistry for work on GPCRs that includes solving the first structures of a ligand-activated GPCR (2r4r, 2r4s, & 2rh1 in 2007)[9][10][11] and the first activated GPCR in complex with its G protein (3sn6 in 2011)[12][13][14][15]. A detailed description of the laureates' body of work on this class of receptors with images is here.
References and Notes
- ↑ Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat Rev Drug Discov. 2006 Dec;5(12):993-6. PMID:17139284 doi:10.1038/nrd2199
- ↑ Peeters MC, van Westen GJ, Li Q, IJzerman AP. Importance of the extracellular loops in G protein-coupled receptors for ligand recognition and receptor activation. Trends Pharmacol Sci. 2011 Jan;32(1):35-42. PMID:21075459 doi:10.1016/j.tips.2010.10.001
- ↑ Millar RP, Newton CL. The year in G protein-coupled receptor research. Mol Endocrinol. 2010 Jan;24(1):261-74. Epub 2009 Dec 17. PMID:20019124 doi:10.1210/me.2009-0473
- ↑ Kristiansen K. Molecular mechanisms of ligand binding, signaling, and regulation within the superfamily of G-protein-coupled receptors: molecular modeling and mutagenesis approaches to receptor structure and function. Pharmacol Ther. 2004 Jul;103(1):21-80. PMID:15251227 doi:10.1016/j.pharmthera.2004.05.002
- ↑ Feng Y, He X, Yang Y, Chao D, Lazarus LH, Xia Y. Current research on opioid receptor function. Curr Drug Targets. 2012 Feb;13(2):230-46. PMID:22204322
- ↑ Donica CL, Awwad HO, Thakker DR, Standifer KM. Cellular mechanisms of nociceptin/orphanin FQ (N/OFQ) peptide (NOP) receptor regulation and heterologous regulation by N/OFQ. Mol Pharmacol. 2013 May;83(5):907-18. doi: 10.1124/mol.112.084632. Epub 2013 Feb , 8. PMID:23395957 doi:http://dx.doi.org/10.1124/mol.112.084632
- ↑ Girault JA, Greengard P. The neurobiology of dopamine signaling. Arch Neurol. 2004 May;61(5):641-4. PMID:15148138 doi:10.1001/archneur.61.5.641
- ↑ Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, Hansen KB, Yuan H, Myers SJ, Dingledine R. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev. 2010 Sep;62(3):405-96. doi: 10.1124/pr.109.002451. PMID:20716669 doi:http://dx.doi.org/10.1124/pr.109.002451
- ↑ Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, Stevens RC. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007 Nov 23;318(5854):1258-65. Epub 2007 Oct 25. PMID:17962520
- ↑ Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Yao XJ, Weis WI, Stevens RC, Kobilka BK. GPCR engineering yields high-resolution structural insights into beta2-adrenergic receptor function. Science. 2007 Nov 23;318(5854):1266-73. Epub 2007 Oct 25. PMID:17962519
- ↑ Ranganathan R. Biochemistry. Signaling across the cell membrane. Science. 2007 Nov 23;318(5854):1253-4. PMID:18033872 doi:10.1126/science.1151656
- ↑ Schwartz TW, Sakmar TP. Structural biology: snapshot of a signalling complex. Nature. 2011 Sep 28;477(7366):540-1. doi: 10.1038/477540a. PMID:21956322 doi:10.1038/477540a
- ↑ Rasmussen SG, DeVree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, Thian FS, Chae PS, Pardon E, Calinski D, Mathiesen JM, Shah ST, Lyons JA, Caffrey M, Gellman SH, Steyaert J, Skiniotis G, Weis WI, Sunahara RK, Kobilka BK. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature. 2011 Jul 19;477(7366):549-55. doi: 10.1038/nature10361. PMID:21772288 doi:10.1038/nature10361
- ↑ Chung KY, Rasmussen SG, Liu T, Li S, DeVree BT, Chae PS, Calinski D, Kobilka BK, Woods VL Jr, Sunahara RK. Conformational changes in the G protein Gs induced by the beta2 adrenergic receptor. Nature. 2011 Sep 28;477(7366):611-5. doi: 10.1038/nature10488. PMID:21956331 doi:10.1038/nature10488
- ↑ Schwartz TW, Sakmar TP. Structural biology: snapshot of a signalling complex. Nature. 2011 Sep 28;477(7366):540-1. doi: 10.1038/477540a. PMID:21956322 doi:10.1038/477540a
See Also
- Nobel Prizes for 3D Molecular Structure
- Highest impact structures of all time
- G proteins
- Rhodopsin
- GTP-binding protein
- Pharmaceutical Drugs
- Membrane proteins
- Hormone
- Ligand Binding N-Terminal of Metabotropic Glutamate Receptors
- Receptor.
Additional Literature
- Carpenter B, Tate CG. Active state structures of G protein-coupled receptors highlight the similarities and differences in the G protein and arrestin coupling interfaces. Curr Opin Struct Biol. 2017 May 5;45:124-132. doi: 10.1016/j.sbi.2017.04.010. PMID:28482214 doi:http://dx.doi.org/10.1016/j.sbi.2017.04.010
External Resources
- Robert J. Lefkowitz and Brian K. Kobilka share the 2012 Nobel Prize in Chemistry for work on GPCRs that includes solving the first structures of a ligand-activated GPCR (2007) and the first activated GPCR in complex with its G protein (2011). A detailed description of the laureates' body of work on this class of receptors with images is here.
- The April 2008 RCSB PDB Molecule of the Month feature on Adrenergic Receptors by David S. Goodsell is 10.2210/rcsb_pdb/mom_2008_4.
- GPCRDB: database contains sequences, ligand binding constants and mutations, in addition GPCR multiple sequence alignments and homology models. Moreover, the site contains useful files where lysozyme and other inserts which are commonly used in the difficult process of crystallizing these transmembrane structures have been removed from the structures.
- GPCR Network site with tracking chart of ongoing structural programs
- The GPCR-SSFE Database: A Homology Model Resource for G-Protein Coupled Receptors
- The blog of the Computational Chemical Biology group at the EMBL-EBI does an excellent job tracking the new GPCR structures as they are emerging.
- Emerald Biosystems Blog that features solved structures and on another page features techniques and amounts needed for crystallization of a number of them.
- A 2012 article from Scripps Research Institute that covers a lot of history of solving the structures of GPCRs and their importance.
- A 2012 article from the Protein Structure Initiative on the screening technique to identify stabilizing fusion partners for solving GPCR structure.
- A 2011 article in Nature entitiled 'Cell Signalling Caught in the Act' describing the first determination of an activated GPCR — the β2 adrenergic receptor (β2AR) — in a complex with its G protein.
- tinyGRAP GPCR mutant database
- GPCR-OKB: GPCR Oligomerization Knowledge Base
- GPCR Natural Variants Database (NaVa)
- The PRED-GPCR server for GPCR recognition and family classification.
- GLASS database: a comprehensive database for experimentally validated GPCR-ligand associations
- GPCR-ModSim is a webserver for computational modeling and simulation of G-Protein Coupled Receptors (GPCRs). Models from sequence and also lets you place the models in a phospholipid bilayer and model them using Molecular Dynamics.
Proteopedia Page Contributors and Editors (what is this?)
Alexander Berchansky, Wayne Decatur, Michal Harel, Ann Taylor, Nikki Hunter